You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001159_01265
You are here: Home > Sequence: MGYG000001159_01265
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bifidobacterium pullorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Bifidobacteriaceae; Bifidobacterium; Bifidobacterium pullorum | |||||||||||
| CAZyme ID | MGYG000001159_01265 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 29323; End: 29931 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 1.02e-20 | 53 | 177 | 5 | 113 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 3.19e-16 | 90 | 177 | 1 | 71 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 3.54e-15 | 70 | 177 | 47 | 136 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGM62627.1 | 1.85e-110 | 1 | 192 | 1 | 192 |
| BAQ29203.1 | 1.85e-110 | 1 | 192 | 1 | 192 |
| AGW85589.1 | 2.24e-55 | 44 | 192 | 22 | 172 |
| QSY56935.1 | 4.96e-50 | 6 | 192 | 3 | 197 |
| QAY61971.1 | 4.59e-32 | 61 | 192 | 528 | 651 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FHE_A | 1.77e-16 | 43 | 183 | 11 | 141 | Highlyactive enzymes by automated modular backbone assembly and sequence design [synthetic construct] |
| 6FHF_A | 7.82e-14 | 61 | 179 | 29 | 141 | Highlyactive enzymes by automated modular backbone assembly and sequence design [Escherichia coli] |
| 2F8Q_A | 6.62e-13 | 61 | 177 | 21 | 134 | Analkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27],2F8Q_B An alkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27] |
| 2FGL_A | 6.64e-13 | 61 | 177 | 22 | 135 | Analkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27],2FGL_B An alkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27] |
| 5EB8_A | 6.67e-13 | 61 | 177 | 23 | 136 | Crystalstructure of aromatic mutant (F4W) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 [Bacillus sp. NG-27],5EB8_B Crystal structure of aromatic mutant (F4W) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 [Bacillus sp. NG-27] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P26223 | 1.13e-14 | 49 | 183 | 3 | 138 | Endo-1,4-beta-xylanase B OS=Butyrivibrio fibrisolvens OX=831 GN=xynB PE=3 SV=1 |
| P10474 | 7.46e-12 | 19 | 139 | 14 | 138 | Endoglucanase/exoglucanase B OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=celB PE=3 SV=1 |
| P38535 | 4.64e-11 | 43 | 184 | 208 | 334 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
| P23557 | 6.72e-11 | 112 | 177 | 31 | 86 | Putative endo-1,4-beta-xylanase OS=Caldicellulosiruptor saccharolyticus OX=44001 PE=3 SV=1 |
| P40944 | 7.84e-11 | 43 | 175 | 349 | 474 | Endo-1,4-beta-xylanase A OS=Caldicellulosiruptor sp. (strain Rt8B.4) OX=28238 GN=xynA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
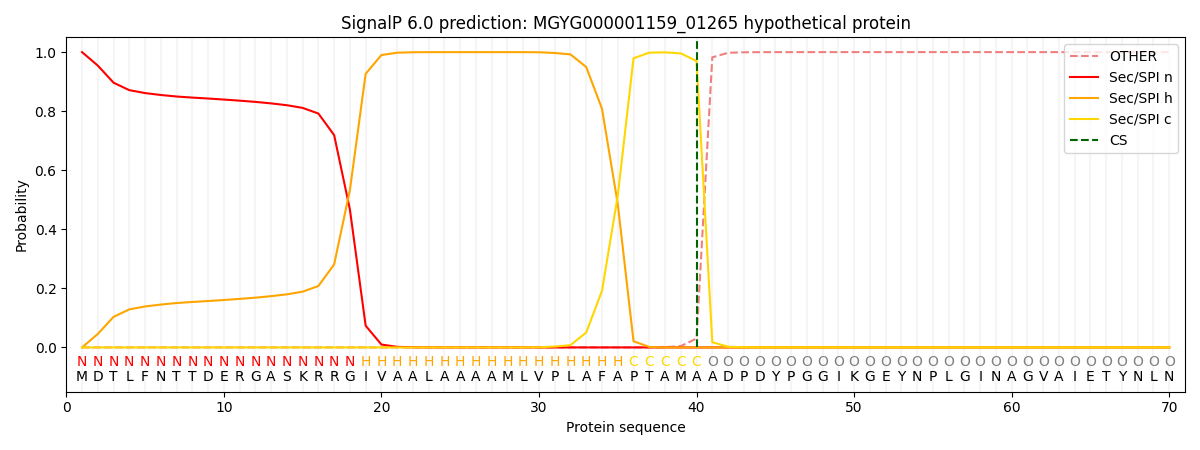
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000840 | 0.997954 | 0.000258 | 0.000410 | 0.000289 | 0.000221 |

