You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001120_00503
You are here: Home > Sequence: MGYG000001120_00503
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Butyricicoccus_A sp002395695 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Butyricicoccaceae; Butyricicoccus_A; Butyricicoccus_A sp002395695 | |||||||||||
| CAZyme ID | MGYG000001120_00503 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 28069; End: 30516 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 164 | 522 | 4.2e-58 | 0.7661538461538462 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 1.70e-58 | 47 | 489 | 42 | 413 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam00295 | Glyco_hydro_28 | 2.22e-21 | 313 | 581 | 85 | 315 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| pfam00395 | SLH | 8.50e-12 | 700 | 741 | 1 | 42 | S-layer homology domain. |
| NF033190 | inl_like_NEAT_1 | 3.78e-10 | 645 | 746 | 644 | 749 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| PLN02218 | PLN02218 | 1.28e-09 | 313 | 525 | 192 | 385 | polygalacturonase ADPG |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AGA79327.1 | 1.73e-95 | 129 | 584 | 74 | 459 |
| QDH80204.1 | 1.34e-93 | 129 | 544 | 74 | 434 |
| AWW33296.1 | 2.62e-93 | 86 | 544 | 53 | 434 |
| QDZ82824.1 | 4.51e-91 | 128 | 582 | 33 | 416 |
| AEN90238.1 | 1.58e-90 | 128 | 582 | 64 | 447 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3JUR_A | 2.08e-66 | 89 | 562 | 26 | 421 | Thecrystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_B The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_C The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_D The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima] |
| 5OLP_A | 8.81e-59 | 99 | 524 | 33 | 402 | Galacturonidase[Bacteroides thetaiotaomicron VPI-5482],5OLP_B Galacturonidase [Bacteroides thetaiotaomicron VPI-5482] |
| 2UVE_A | 8.22e-12 | 128 | 521 | 172 | 530 | Structureof Yersinia enterocolitica Family 28 Exopolygalacturonase [Yersinia enterocolitica],2UVE_B Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase [Yersinia enterocolitica],2UVF_A Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase in Complex with Digalaturonic Acid [Yersinia enterocolitica],2UVF_B Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase in Complex with Digalaturonic Acid [Yersinia enterocolitica] |
| 1NHC_A | 2.75e-06 | 281 | 488 | 65 | 266 | Structuralinsights into the processivity of endopolygalacturonase I from Aspergillus niger [Aspergillus niger],1NHC_B Structural insights into the processivity of endopolygalacturonase I from Aspergillus niger [Aspergillus niger],1NHC_C Structural insights into the processivity of endopolygalacturonase I from Aspergillus niger [Aspergillus niger],1NHC_D Structural insights into the processivity of endopolygalacturonase I from Aspergillus niger [Aspergillus niger],1NHC_E Structural insights into the processivity of endopolygalacturonase I from Aspergillus niger [Aspergillus niger],1NHC_F Structural insights into the processivity of endopolygalacturonase I from Aspergillus niger [Aspergillus niger] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P38537 | 3.31e-17 | 651 | 811 | 45 | 203 | Surface-layer 125 kDa protein OS=Lysinibacillus sphaericus OX=1421 PE=3 SV=1 |
| A7PZL3 | 7.92e-15 | 128 | 552 | 78 | 451 | Probable polygalacturonase OS=Vitis vinifera OX=29760 GN=GSVIVT00026920001 PE=1 SV=1 |
| P15922 | 2.57e-11 | 67 | 517 | 129 | 524 | Exo-poly-alpha-D-galacturonosidase OS=Dickeya chrysanthemi OX=556 GN=pehX PE=1 SV=1 |
| P19424 | 5.90e-11 | 640 | 813 | 41 | 213 | Endoglucanase OS=Bacillus sp. (strain KSM-635) OX=1415 PE=1 SV=1 |
| P38536 | 3.65e-10 | 623 | 813 | 1665 | 1854 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
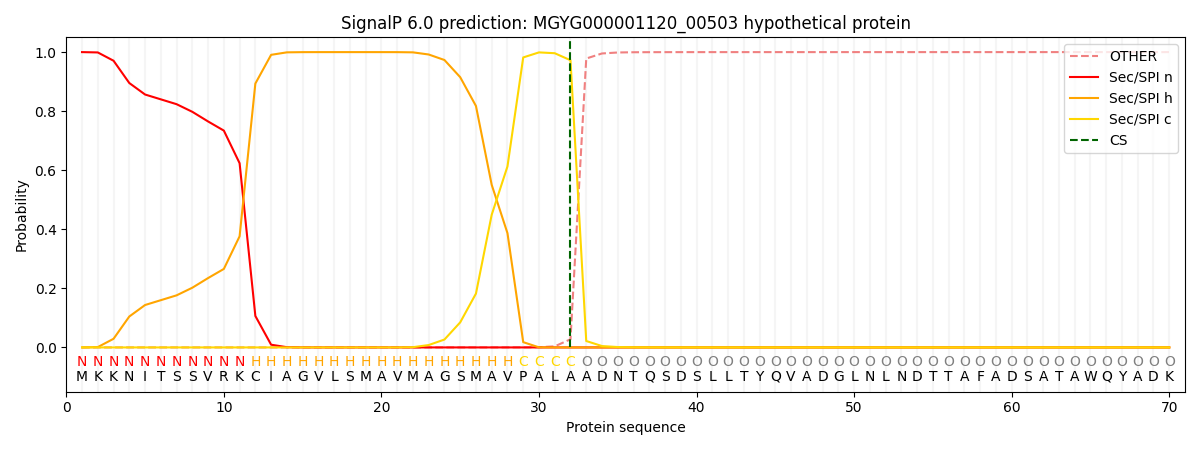
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000302 | 0.998901 | 0.000186 | 0.000232 | 0.000192 | 0.000161 |

