You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001065_01643
You are here: Home > Sequence: MGYG000001065_01643
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
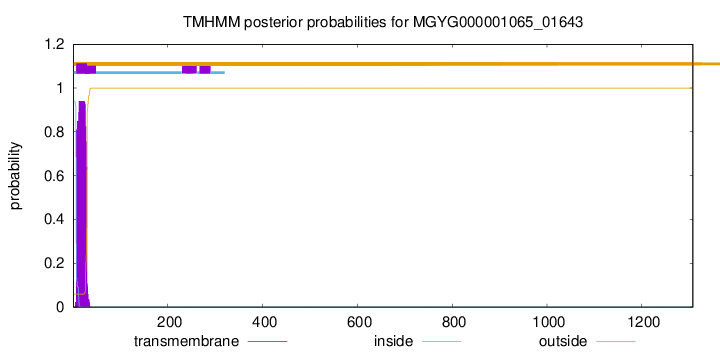
TMHMM annotations
Basic Information help
| Species | Enterocloster sp900538485 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Enterocloster; Enterocloster sp900538485 | |||||||||||
| CAZyme ID | MGYG000001065_01643 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10462; End: 14388 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 275 | 686 | 6.4e-54 | 0.9323076923076923 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 8.74e-64 | 121 | 667 | 3 | 472 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| NF033840 | PspC_relate_1 | 9.37e-10 | 1196 | 1306 | 518 | 646 | PspC-related protein choline-binding protein 1. Members of this family share C-terminal homology to the choline-binding form of the pneumococcal surface antigen PspC, but not to its allelic LPXTG-anchored forms because they lack the choline-binding repeat region. Members of this family should not be confused with PspC itself, whose identity and function reflect regions N-terminal to the choline-binding region. See Iannelli, et al. (PMID: 11891047) for information about the different allelic forms of PspC. |
| pfam00295 | Glyco_hydro_28 | 1.15e-08 | 442 | 596 | 83 | 229 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| pfam02368 | Big_2 | 1.82e-08 | 1059 | 1124 | 2 | 70 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
| PLN02218 | PLN02218 | 3.28e-08 | 444 | 660 | 192 | 396 | polygalacturonase ADPG |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SET92437.1 | 0.0 | 104 | 1054 | 93 | 1066 |
| QOV20136.1 | 9.31e-268 | 99 | 1051 | 57 | 1070 |
| AQS56465.1 | 1.33e-163 | 118 | 1051 | 24 | 944 |
| AOV08612.1 | 6.29e-163 | 110 | 1051 | 29 | 959 |
| BAB85762.1 | 5.72e-161 | 107 | 1028 | 26 | 927 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2UVE_A | 1.95e-120 | 109 | 702 | 31 | 582 | Structureof Yersinia enterocolitica Family 28 Exopolygalacturonase [Yersinia enterocolitica],2UVE_B Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase [Yersinia enterocolitica],2UVF_A Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase in Complex with Digalaturonic Acid [Yersinia enterocolitica],2UVF_B Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase in Complex with Digalaturonic Acid [Yersinia enterocolitica] |
| 5OLP_A | 5.63e-16 | 238 | 614 | 45 | 368 | Galacturonidase[Bacteroides thetaiotaomicron VPI-5482],5OLP_B Galacturonidase [Bacteroides thetaiotaomicron VPI-5482] |
| 3JUR_A | 3.67e-13 | 233 | 600 | 24 | 345 | Thecrystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_B The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_C The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_D The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima] |
| 1BHE_A | 9.22e-10 | 267 | 616 | 26 | 314 | ChainA, POLYGALACTURONASE [Pectobacterium carotovorum] |
| 4MXN_A | 2.58e-07 | 259 | 363 | 30 | 121 | Crystalstructure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_B Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_C Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_D Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P15922 | 3.34e-119 | 119 | 727 | 36 | 599 | Exo-poly-alpha-D-galacturonosidase OS=Dickeya chrysanthemi OX=556 GN=pehX PE=1 SV=1 |
| P20041 | 1.17e-16 | 270 | 619 | 76 | 402 | Polygalacturonase OS=Ralstonia solanacearum OX=305 GN=pglA PE=1 SV=1 |
| P58598 | 6.36e-16 | 270 | 617 | 78 | 402 | Polygalacturonase OS=Ralstonia solanacearum (strain GMI1000) OX=267608 GN=pglA PE=3 SV=1 |
| P18192 | 2.45e-09 | 267 | 616 | 52 | 340 | Endo-polygalacturonase OS=Pectobacterium carotovorum subsp. carotovorum OX=555 GN=peh PE=3 SV=1 |
| P26509 | 5.71e-09 | 267 | 616 | 52 | 340 | Endo-polygalacturonase OS=Pectobacterium parmentieri OX=1905730 GN=pehA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
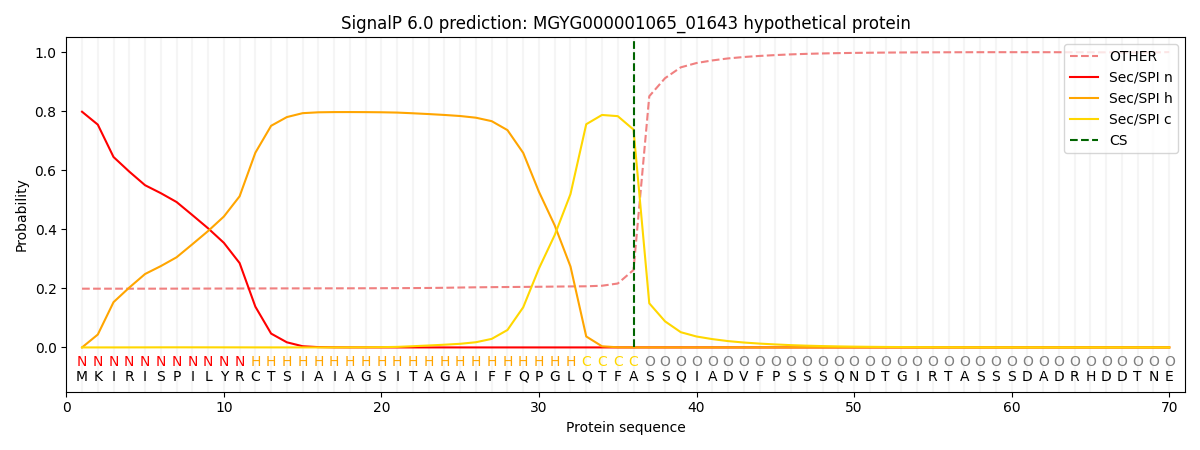
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.212330 | 0.782873 | 0.003580 | 0.000480 | 0.000327 | 0.000396 |