You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000866_00635
You are here: Home > Sequence: MGYG000000866_00635
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
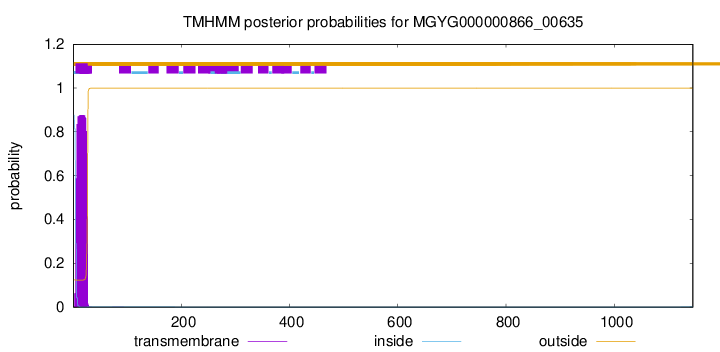
TMHMM annotations
Basic Information help
| Species | Prevotellamassilia sp900556465 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotellamassilia; Prevotellamassilia sp900556465 | |||||||||||
| CAZyme ID | MGYG000000866_00635 | |||||||||||
| CAZy Family | GH33 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5065; End: 8502 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH33 | 383 | 737 | 9e-45 | 0.9269005847953217 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd15482 | Sialidase_non-viral | 1.21e-62 | 372 | 742 | 1 | 339 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| pfam13088 | BNR_2 | 7.57e-09 | 407 | 612 | 10 | 194 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QFQ12979.1 | 1.30e-109 | 372 | 754 | 21 | 400 |
| ALO48969.1 | 1.31e-107 | 358 | 752 | 13 | 400 |
| QYR10380.1 | 1.10e-102 | 374 | 751 | 27 | 395 |
| QUB85277.1 | 6.26e-99 | 256 | 958 | 245 | 984 |
| QUB76590.1 | 6.29e-99 | 283 | 1010 | 294 | 1042 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1EUR_A | 4.15e-16 | 361 | 680 | 1 | 294 | Sialidase[Micromonospora viridifaciens],1EUS_A Sialidase Complexed With 2-Deoxy-2,3-Dehydro-N- Acetylneuraminic Acid [Micromonospora viridifaciens] |
| 1EUT_A | 1.47e-15 | 361 | 680 | 1 | 294 | Sialidase,Large 68kd Form, Complexed With Galactose [Micromonospora viridifaciens],1EUU_A Sialidase Or Neuraminidase, Large 68kd Form [Micromonospora viridifaciens] |
| 7LBU_A | 6.16e-15 | 370 | 598 | 79 | 303 | ChainA, Exo-alpha-sialidase [Cutibacterium acnes],7LBV_A Chain A, Exo-alpha-sialidase [Cutibacterium acnes] |
| 2BER_A | 3.05e-14 | 384 | 680 | 21 | 290 | Y370GActive Site Mutant of the Sialidase from Micromonospora viridifaciens in complex with beta-Neu5Ac (sialic acid). [Micromonospora viridifaciens] |
| 1WCQ_A | 3.05e-14 | 384 | 680 | 21 | 290 | Mutagenesisof the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens],1WCQ_B Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens],1WCQ_C Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q02834 | 8.79e-15 | 361 | 680 | 43 | 336 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
| P15698 | 1.03e-14 | 384 | 604 | 54 | 269 | Sialidase OS=Paeniclostridium sordellii OX=1505 PE=1 SV=1 |
| P10481 | 1.13e-11 | 384 | 598 | 36 | 245 | Sialidase OS=Clostridium perfringens OX=1502 GN=nanH PE=1 SV=1 |
| P62575 | 2.99e-08 | 360 | 638 | 315 | 684 | Sialidase A OS=Streptococcus pneumoniae OX=1313 GN=nanA PE=1 SV=1 |
| P62576 | 2.99e-08 | 360 | 638 | 315 | 684 | Sialidase A OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=nanA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
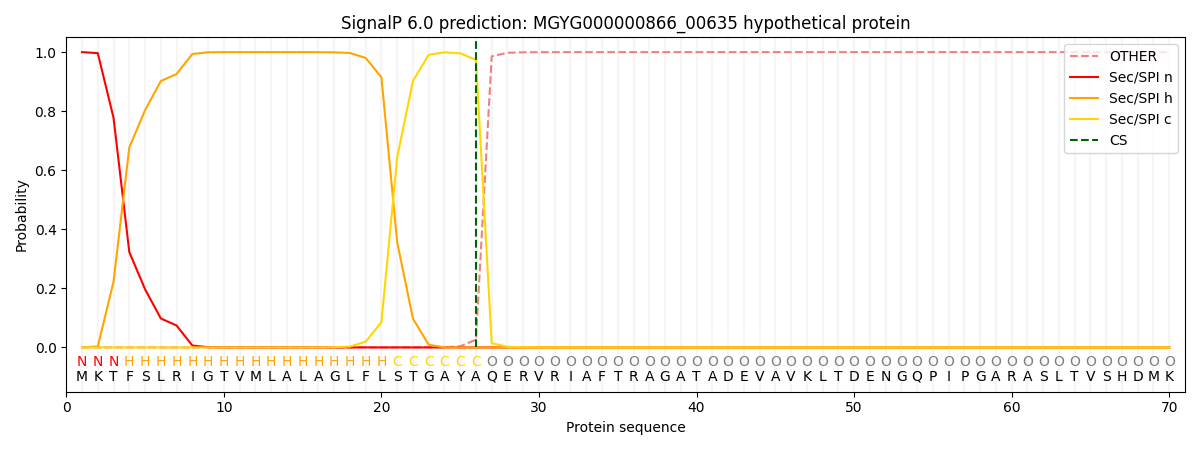
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000851 | 0.998128 | 0.000346 | 0.000260 | 0.000204 | 0.000194 |