You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000720_00067
You are here: Home > Sequence: MGYG000000720_00067
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
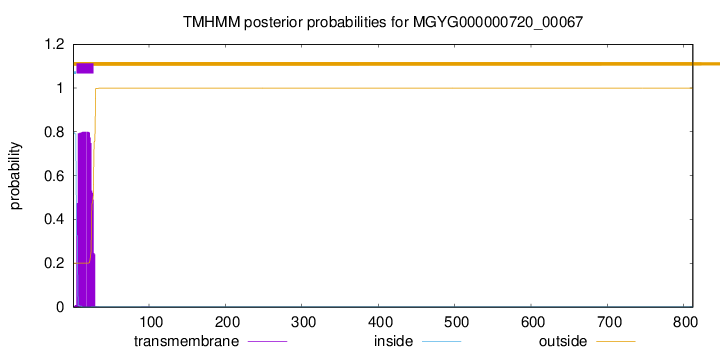
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; | |||||||||||
| CAZyme ID | MGYG000000720_00067 | |||||||||||
| CAZy Family | CBM85 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 67210; End: 69648 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 461 | 750 | 1.5e-50 | 0.9471947194719472 |
| CBM85 | 267 | 396 | 3.9e-20 | 0.9696969696969697 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 7.49e-40 | 523 | 727 | 20 | 235 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 7.81e-32 | 491 | 726 | 58 | 302 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 9.57e-28 | 464 | 726 | 12 | 275 | Glycosyl hydrolase family 10. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDU70528.1 | 3.69e-108 | 262 | 811 | 66 | 612 |
| BAM04877.1 | 5.59e-100 | 269 | 811 | 101 | 638 |
| QDU61652.1 | 1.08e-99 | 270 | 788 | 71 | 591 |
| AHF89846.1 | 1.96e-95 | 303 | 792 | 97 | 595 |
| ACB76489.1 | 1.53e-94 | 237 | 806 | 29 | 598 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7D88_A | 9.29e-36 | 418 | 800 | 42 | 405 | ChainA, Beta-xylanase [Bacillus sp. (in: Bacteria)] |
| 7D89_A | 3.43e-34 | 418 | 800 | 42 | 405 | ChainA, Beta-xylanase [Bacillus sp. (in: Bacteria)] |
| 4W8L_A | 3.05e-16 | 523 | 725 | 66 | 305 | Structureof GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4W8L_B Structure of GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4W8L_C Structure of GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis] |
| 6FHF_A | 1.09e-15 | 491 | 695 | 50 | 270 | Highlyactive enzymes by automated modular backbone assembly and sequence design [Escherichia coli] |
| 3NIY_A | 2.10e-15 | 518 | 726 | 76 | 299 | Crystalstructure of native xylanase 10B from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3NIY_B Crystal structure of native xylanase 10B from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3NJ3_A Crystal structure of xylanase 10B from Thermotoga petrophila RKU-1 in complex with xylobiose [Thermotoga petrophila RKU-1],3NJ3_B Crystal structure of xylanase 10B from Thermotoga petrophila RKU-1 in complex with xylobiose [Thermotoga petrophila RKU-1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A3DH97 | 1.10e-38 | 407 | 812 | 365 | 757 | Anti-sigma-I factor RsgI6 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=rsgI6 PE=1 SV=1 |
| A0A1P8AWH8 | 3.15e-30 | 429 | 798 | 569 | 926 | Endo-1,4-beta-xylanase 1 OS=Arabidopsis thaliana OX=3702 GN=XYN1 PE=1 SV=1 |
| O80596 | 7.69e-29 | 320 | 810 | 604 | 1060 | Endo-1,4-beta-xylanase 2 OS=Arabidopsis thaliana OX=3702 GN=XYN2 PE=3 SV=1 |
| F4JG10 | 5.03e-26 | 428 | 810 | 373 | 745 | Endo-1,4-beta-xylanase 3 OS=Arabidopsis thaliana OX=3702 GN=XYN3 PE=2 SV=1 |
| Q84WT5 | 3.70e-22 | 418 | 798 | 171 | 549 | Endo-1,4-beta-xylanase 5-like OS=Arabidopsis thaliana OX=3702 GN=At4g33820 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
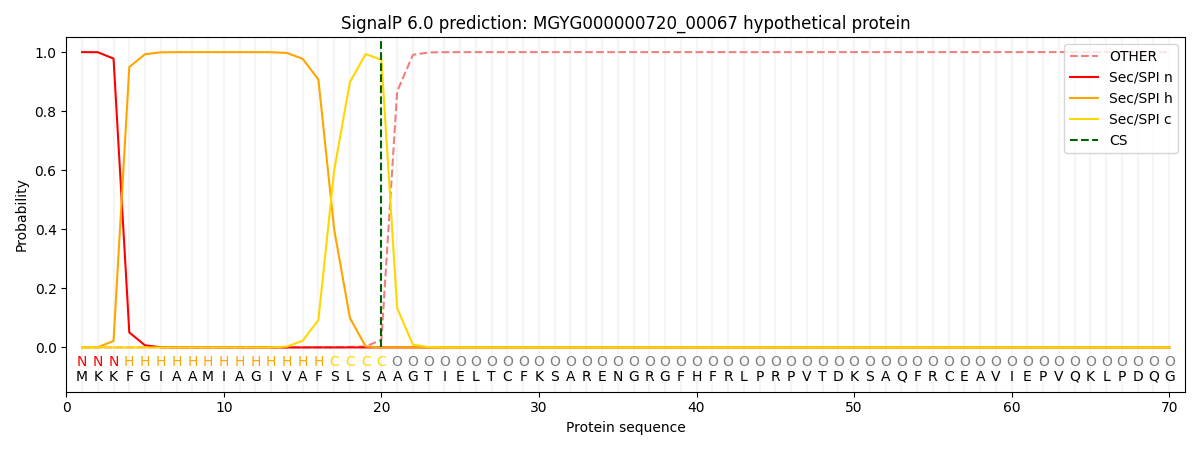
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000285 | 0.999067 | 0.000182 | 0.000146 | 0.000141 | 0.000134 |