You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000695_02848
You are here: Home > Sequence: MGYG000000695_02848
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp000436035 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp000436035 | |||||||||||
| CAZyme ID | MGYG000000695_02848 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4279; End: 6840 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 435 | 649 | 1.6e-42 | 0.7326388888888888 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4677 | PemB | 1.52e-15 | 440 | 631 | 112 | 332 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| PRK10531 | PRK10531 | 5.73e-13 | 410 | 637 | 81 | 354 | putative acyl-CoA thioester hydrolase. |
| pfam01095 | Pectinesterase | 2.64e-12 | 413 | 643 | 2 | 233 | Pectinesterase. |
| PLN02708 | PLN02708 | 1.19e-11 | 434 | 643 | 259 | 484 | Probable pectinesterase/pectinesterase inhibitor |
| PLN02488 | PLN02488 | 2.03e-11 | 414 | 630 | 200 | 417 | probable pectinesterase/pectinesterase inhibitor |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AGB28243.1 | 7.58e-224 | 25 | 853 | 557 | 1371 |
| VEH16322.1 | 6.23e-207 | 28 | 853 | 562 | 1471 |
| QUT74167.1 | 1.04e-87 | 25 | 853 | 401 | 1435 |
| QCD38476.1 | 4.12e-77 | 433 | 853 | 1057 | 1476 |
| QCP72166.1 | 4.12e-77 | 433 | 853 | 1057 | 1476 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1XG2_A | 2.08e-09 | 413 | 635 | 5 | 228 | ChainA, Pectinesterase 1 [Solanum lycopersicum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q1PEC0 | 2.19e-10 | 414 | 643 | 217 | 445 | Probable pectinesterase/pectinesterase inhibitor 42 OS=Arabidopsis thaliana OX=3702 GN=PME42 PE=2 SV=1 |
| Q9M9W7 | 3.00e-10 | 414 | 645 | 242 | 465 | Putative pectinesterase/pectinesterase inhibitor 22 OS=Arabidopsis thaliana OX=3702 GN=PME22 PE=3 SV=1 |
| O81320 | 5.92e-10 | 414 | 650 | 166 | 402 | Putative pectinesterase/pectinesterase inhibitor 38 OS=Arabidopsis thaliana OX=3702 GN=PME38 PE=3 SV=1 |
| P09607 | 8.38e-09 | 413 | 584 | 238 | 402 | Pectinesterase 2.1 OS=Solanum lycopersicum OX=4081 GN=PME2.1 PE=2 SV=2 |
| Q1JPL7 | 8.47e-09 | 410 | 644 | 246 | 479 | Pectinesterase/pectinesterase inhibitor 18 OS=Arabidopsis thaliana OX=3702 GN=PME18 PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP
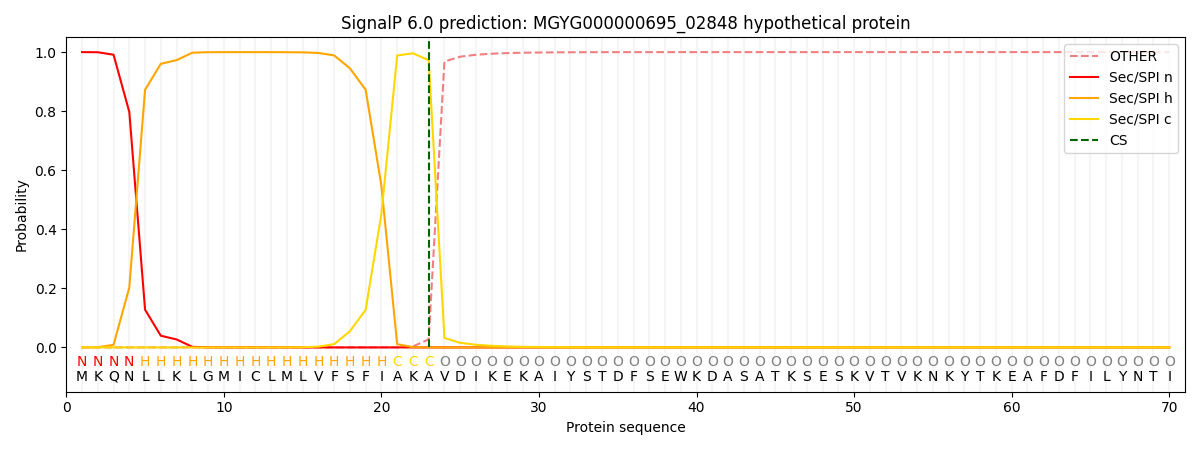
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000907 | 0.998112 | 0.000275 | 0.000234 | 0.000213 | 0.000206 |
