You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000587_00513
You are here: Home > Sequence: MGYG000000587_00513
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales_A; UBA1381; ; | |||||||||||
| CAZyme ID | MGYG000000587_00513 | |||||||||||
| CAZy Family | GH141 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1611; End: 5162 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH141 | 36 | 549 | 9.9e-134 | 0.9620493358633776 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd10238 | HSPA14-like_NBD | 7.42e-04 | 688 | 816 | 1 | 128 | Nucleotide-binding domain of human HSPA14 and similar proteins. Human HSPA14 (also known as 70-kDa heat shock protein 14, HSP70L1, HSP70-4; the gene encoding HSPA14 maps to 10p13), is ribosome-associated and belongs to the heat shock protein 70 (HSP70) family of chaperones that assist in protein folding and assembly, and can direct incompetent "client" proteins towards degradation. Typically, HSP70s have a nucleotide-binding domain (NBD) and a substrate-binding domain (SBD). The nucleotide sits in a deep cleft formed between the two lobes of the NBD. The two subdomains of each lobe change conformation between ATP-bound, ADP-bound, and nucleotide-free states. ATP binding opens up the substrate-binding site; substrate-binding increases the rate of ATP hydrolysis. HSP70 chaperone activity is regulated by various co-chaperones: J-domain proteins and nucleotide exchange factors (NEFs). HSPA14 interacts with the J-protein MPP11 to form the mammalian ribosome-associated complex (mRAC). HSPA14 participates in a pathway along with Nijmegen breakage syndrome 1 (NBS1, also known as p85 or nibrin), heat shock transcription factor 4b (HSF4b), and HSPA4 (belonging to a different subfamily), that induces tumor migration, invasion, and transformation. HSPA14 is a potent T helper cell (Th1) polarizing adjuvant that contributes to antitumor immune responses. |
| pfam00395 | SLH | 0.001 | 1130 | 1172 | 1 | 42 | S-layer homology domain. |
| pfam06439 | DUF1080 | 0.006 | 824 | 991 | 4 | 167 | Domain of Unknown Function (DUF1080). This family has structural similarity to an endo-1,3-1,4-beta glucanase belonging to glycoside hydrolase family 16. However, the structure surrounding the active site differs from that of the endo-1,3-1,4-beta glucanase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUL54275.1 | 3.40e-141 | 24 | 992 | 395 | 1348 |
| AIQ43746.1 | 4.64e-140 | 24 | 992 | 395 | 1348 |
| QYR21751.1 | 2.33e-138 | 39 | 1018 | 426 | 1384 |
| ANE49726.1 | 4.36e-102 | 3 | 623 | 11 | 628 |
| SDS22682.1 | 2.57e-92 | 28 | 623 | 41 | 606 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQP_A | 5.33e-47 | 19 | 539 | 29 | 574 | Glycosidehydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_B Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_C Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_D Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_E Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_F Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_G Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_H Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
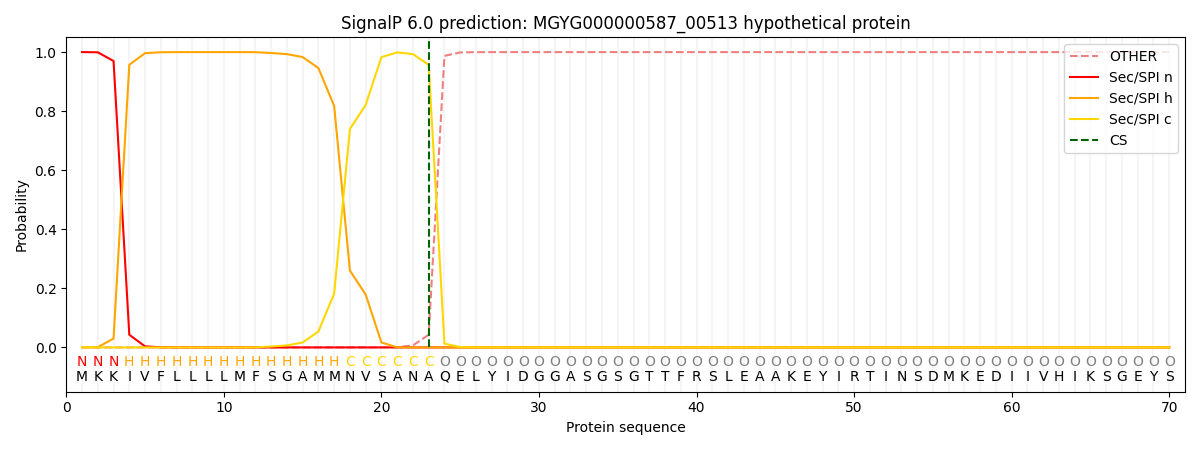
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000306 | 0.998975 | 0.000233 | 0.000166 | 0.000158 | 0.000152 |
