You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000555_00487
You are here: Home > Sequence: MGYG000000555_00487
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cellulosilyticum sp900556665 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Cellulosilyticaceae; Cellulosilyticum; Cellulosilyticum sp900556665 | |||||||||||
| CAZyme ID | MGYG000000555_00487 | |||||||||||
| CAZy Family | GH14 | |||||||||||
| CAZyme Description | Thermophilic beta-amylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12005; End: 13732 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH14 | 41 | 455 | 5.6e-116 | 0.970873786407767 |
| CBM20 | 479 | 564 | 5.2e-30 | 0.9333333333333333 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01373 | Glyco_hydro_14 | 1.25e-118 | 41 | 454 | 3 | 390 | Glycosyl hydrolase family 14. This family are beta amylases. |
| PLN02801 | PLN02801 | 5.26e-64 | 41 | 468 | 21 | 444 | beta-amylase |
| PLN00197 | PLN00197 | 1.18e-61 | 13 | 428 | 100 | 488 | beta-amylase; Provisional |
| PLN02803 | PLN02803 | 4.15e-56 | 17 | 483 | 84 | 533 | beta-amylase |
| PLN02905 | PLN02905 | 1.44e-49 | 41 | 472 | 270 | 695 | beta-amylase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ACZ98633.1 | 8.04e-175 | 1 | 300 | 1 | 297 |
| ACZ98658.1 | 2.47e-166 | 306 | 574 | 1 | 269 |
| AJQ93792.1 | 4.63e-165 | 37 | 558 | 29 | 547 |
| QKE84418.1 | 4.01e-150 | 36 | 474 | 47 | 489 |
| ADB75269.1 | 7.71e-150 | 2 | 469 | 4 | 475 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1VEP_A | 5.20e-105 | 31 | 553 | 6 | 496 | ChainA, Beta-amylase [Bacillus cereus] |
| 1VEN_A | 7.33e-105 | 31 | 553 | 6 | 496 | ChainA, Beta-amylase [Bacillus cereus] |
| 1B90_A | 1.46e-104 | 31 | 553 | 6 | 496 | BacillusCereus Beta-Amylase Apo Form [Bacillus cereus],1B9Z_A Bacillus Cereus Beta-Amylase Complexed With Maltose [Bacillus cereus],1J0Y_A Beta-amylase from Bacillus cereus var. mycoides in complex with glucose [Bacillus cereus],1J0Y_B Beta-amylase from Bacillus cereus var. mycoides in complex with glucose [Bacillus cereus],1J0Y_C Beta-amylase from Bacillus cereus var. mycoides in complex with glucose [Bacillus cereus],1J0Y_D Beta-amylase from Bacillus cereus var. mycoides in complex with glucose [Bacillus cereus],1J0Z_A Beta-amylase from Bacillus cereus var. mycoides in complex with maltose [Bacillus cereus],1J0Z_B Beta-amylase from Bacillus cereus var. mycoides in complex with maltose [Bacillus cereus],1J0Z_C Beta-amylase from Bacillus cereus var. mycoides in complex with maltose [Bacillus cereus],1J0Z_D Beta-amylase from Bacillus cereus var. mycoides in complex with maltose [Bacillus cereus],1J10_A beta-amylase from Bacillus cereus var. mycoides in complex with GGX [Bacillus cereus],1J10_B beta-amylase from Bacillus cereus var. mycoides in complex with GGX [Bacillus cereus],1J10_C beta-amylase from Bacillus cereus var. mycoides in complex with GGX [Bacillus cereus],1J10_D beta-amylase from Bacillus cereus var. mycoides in complex with GGX [Bacillus cereus],1J11_A beta-amylase from Bacillus cereus var. mycoides in complex with alpha-EPG [Bacillus cereus],1J11_B beta-amylase from Bacillus cereus var. mycoides in complex with alpha-EPG [Bacillus cereus],1J11_C beta-amylase from Bacillus cereus var. mycoides in complex with alpha-EPG [Bacillus cereus],1J11_D beta-amylase from Bacillus cereus var. mycoides in complex with alpha-EPG [Bacillus cereus],1J12_A Beta-Amylase from Bacillus cereus var. mycoides in Complex with alpha-EBG [Bacillus cereus],1J12_B Beta-Amylase from Bacillus cereus var. mycoides in Complex with alpha-EBG [Bacillus cereus],1J12_C Beta-Amylase from Bacillus cereus var. mycoides in Complex with alpha-EBG [Bacillus cereus],1J12_D Beta-Amylase from Bacillus cereus var. mycoides in Complex with alpha-EBG [Bacillus cereus],1J18_A Crystal Structure of a Beta-Amylase from Bacillus cereus var. mycoides Cocrystallized with Maltose [Bacillus cereus],1VEM_A Crystal Structure Analysis of Bacillus Cereus Beta-Amylase at the optimum pH (6.5) [Bacillus cereus],5BCA_A Beta-Amylase From Bacillus Cereus Var. Mycoides [Bacillus cereus],5BCA_B Beta-Amylase From Bacillus Cereus Var. Mycoides [Bacillus cereus],5BCA_C Beta-Amylase From Bacillus Cereus Var. Mycoides [Bacillus cereus],5BCA_D Beta-Amylase From Bacillus Cereus Var. Mycoides [Bacillus cereus] |
| 1VEO_A | 2.05e-104 | 31 | 553 | 6 | 496 | ChainA, Beta-amylase [Bacillus cereus] |
| 1ITC_A | 1.14e-103 | 31 | 553 | 6 | 496 | ChainA, Beta-Amylase [Bacillus cereus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P19584 | 1.53e-120 | 4 | 560 | 6 | 535 | Thermophilic beta-amylase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 PE=1 SV=1 |
| P36924 | 1.89e-103 | 31 | 553 | 36 | 526 | Beta-amylase OS=Bacillus cereus OX=1396 GN=spoII PE=1 SV=2 |
| P06547 | 7.57e-92 | 4 | 570 | 6 | 544 | Beta-amylase OS=Niallia circulans OX=1397 PE=3 SV=1 |
| P96513 | 1.71e-87 | 35 | 475 | 40 | 454 | Beta-amylase (Fragment) OS=Cytobacillus firmus OX=1399 PE=3 SV=1 |
| P21543 | 1.19e-84 | 35 | 569 | 40 | 540 | Beta/alpha-amylase OS=Paenibacillus polymyxa OX=1406 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
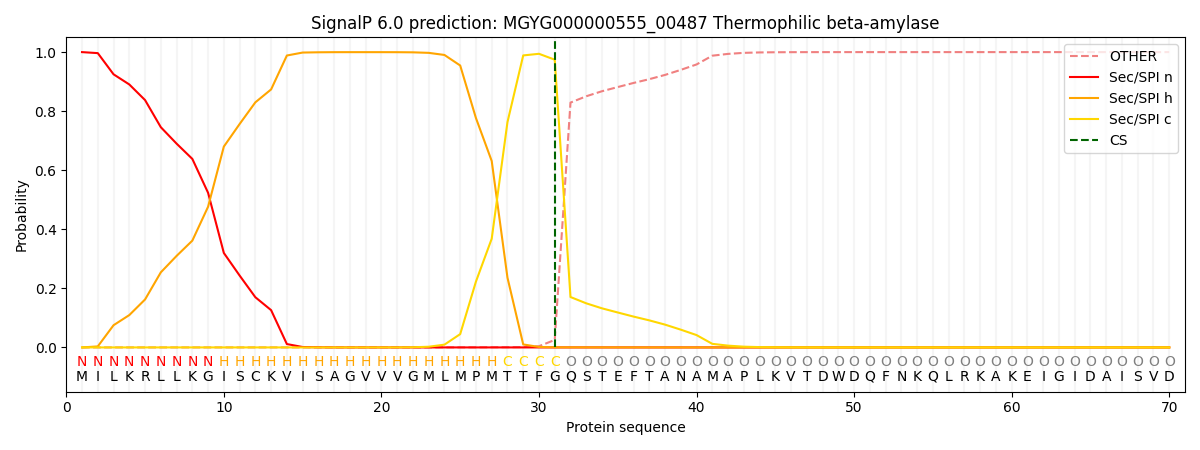
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000476 | 0.998774 | 0.000192 | 0.000214 | 0.000172 | 0.000151 |
