You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000442_01835
You are here: Home > Sequence: MGYG000000442_01835
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-462 sp003489705 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; CAG-462; CAG-462 sp003489705 | |||||||||||
| CAZyme ID | MGYG000000442_01835 | |||||||||||
| CAZy Family | GH105 | |||||||||||
| CAZyme Description | Unsaturated rhamnogalacturonyl hydrolase YteR | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 31410; End: 32624 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH105 | 54 | 397 | 1.6e-117 | 0.9789156626506024 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07470 | Glyco_hydro_88 | 2.75e-118 | 34 | 399 | 1 | 343 | Glycosyl Hydrolase Family 88. Unsaturated glucuronyl hydrolase catalyzes the hydrolytic release of unsaturated glucuronic acids from oligosaccharides (EC:3.2.1.-) produced by the reactions of polysaccharide lyases. |
| COG4225 | YesR | 4.03e-107 | 60 | 398 | 34 | 356 | Rhamnogalacturonyl hydrolase YesR [Carbohydrate transport and metabolism]. |
| pfam05147 | LANC_like | 2.24e-04 | 65 | 346 | 13 | 266 | Lanthionine synthetase C-like protein. Lanthionines are thioether bridges that are putatively generated by dehydration of Ser and Thr residues followed by addition of cysteine residues within the peptide. This family contains the lanthionine synthetase C-like proteins 1 and 2 which are related to the bacterial lanthionine synthetase components C (LanC). LANCL1 (P40 seven-transmembrane-domain protein) and LANCL2 (testes-specific adriamycin sensitivity protein) are thought to be peptide-modifying enzyme components in eukaryotic cells. Both proteins are produced in large quantities in the brain and testes and may have role in the immune surveillance of these organs. Lanthionines are found in lantibiotics, which are peptide-derived, post-translationally modified antimicrobials produced by several bacterial strains. This region contains seven internal repeats. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SCV08369.1 | 7.84e-186 | 12 | 399 | 11 | 398 |
| QUR41787.1 | 7.84e-186 | 12 | 399 | 11 | 398 |
| QRQ55761.1 | 7.84e-186 | 12 | 399 | 11 | 398 |
| QUT32871.1 | 7.84e-186 | 12 | 399 | 11 | 398 |
| ALJ48950.1 | 7.84e-186 | 12 | 399 | 11 | 398 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4WU0_A | 1.01e-111 | 60 | 398 | 23 | 361 | StructuralAnalysis of C. acetobutylicum ATCC 824 Glycoside Hydrolase From Family 105 [Clostridium acetobutylicum ATCC 824],4WU0_B Structural Analysis of C. acetobutylicum ATCC 824 Glycoside Hydrolase From Family 105 [Clostridium acetobutylicum ATCC 824] |
| 1NC5_A | 9.29e-88 | 59 | 404 | 38 | 373 | Structureof Protein of Unknown Function of YteR from Bacillus Subtilis [Bacillus subtilis],2D8L_A Crystal Structure of Unsaturated Rhamnogalacturonyl Hydrolase in complex with dGlcA-GalNAc [Bacillus subtilis] |
| 2GH4_A | 3.85e-87 | 59 | 404 | 28 | 363 | ChainA, Putative glycosyl hydrolase yteR [Bacillus subtilis] |
| 3QWT_A | 6.30e-21 | 60 | 336 | 65 | 329 | ChainA, Putative GH105 family protein [Salmonella enterica subsp. enterica serovar Paratyphi A],3QWT_B Chain B, Putative GH105 family protein [Salmonella enterica subsp. enterica serovar Paratyphi A],3QWT_C Chain C, Putative GH105 family protein [Salmonella enterica subsp. enterica serovar Paratyphi A],3QWT_D Chain D, Putative GH105 family protein [Salmonella enterica subsp. enterica serovar Paratyphi A] |
| 3K11_A | 8.03e-16 | 62 | 398 | 82 | 413 | Crystalstructure of Putative glycosyl hydrolase (NP_813087.1) from BACTEROIDES THETAIOTAOMICRON VPI-5482 at 1.80 A resolution [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34559 | 5.09e-87 | 59 | 404 | 38 | 373 | Unsaturated rhamnogalacturonyl hydrolase YteR OS=Bacillus subtilis (strain 168) OX=224308 GN=yteR PE=1 SV=1 |
| P0A3U6 | 4.12e-48 | 171 | 394 | 1 | 227 | Protein Atu3128 OS=Agrobacterium fabrum (strain C58 / ATCC 33970) OX=176299 GN=Atu3128 PE=3 SV=1 |
| P0A3U7 | 4.12e-48 | 171 | 394 | 1 | 227 | 24.9 kDa protein in picA locus OS=Rhizobium radiobacter OX=358 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
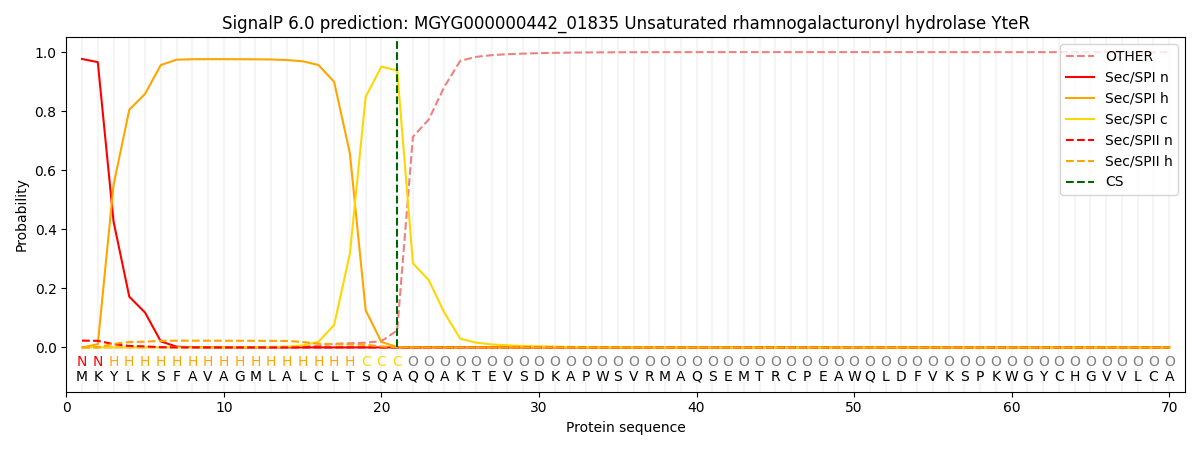
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001046 | 0.973256 | 0.024781 | 0.000390 | 0.000268 | 0.000235 |
