You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000345_02831
You are here: Home > Sequence: MGYG000000345_02831
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA1829 sp002338895 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; UBA1829; UBA1829 sp002338895 | |||||||||||
| CAZyme ID | MGYG000000345_02831 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 29769; End: 32948 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 367 | 539 | 2.8e-24 | 0.39211136890951276 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3693 | XynA | 5.23e-13 | 312 | 522 | 16 | 222 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam01229 | Glyco_hydro_39 | 9.74e-09 | 342 | 529 | 42 | 227 | Glycosyl hydrolases family 39. |
| smart00633 | Glyco_10 | 5.50e-07 | 361 | 454 | 3 | 86 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 3.08e-06 | 361 | 494 | 46 | 167 | Glycosyl hydrolase family 10. |
| pfam02449 | Glyco_hydro_42 | 9.60e-06 | 338 | 457 | 14 | 141 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44457.1 | 0.0 | 31 | 1052 | 32 | 1061 |
| AVM45381.1 | 4.16e-156 | 30 | 1052 | 30 | 1042 |
| QIZ71564.1 | 1.89e-22 | 352 | 592 | 72 | 319 |
| AFY82267.1 | 1.66e-21 | 356 | 557 | 85 | 283 |
| QIZ71566.1 | 2.59e-21 | 352 | 591 | 77 | 316 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4UCF_A | 4.86e-08 | 338 | 457 | 38 | 165 | Crystalstructure of Bifidobacterium bifidum beta-galactosidase in complex with alpha-galactose [Bifidobacterium bifidum S17],4UCF_B Crystal structure of Bifidobacterium bifidum beta-galactosidase in complex with alpha-galactose [Bifidobacterium bifidum S17],4UCF_C Crystal structure of Bifidobacterium bifidum beta-galactosidase in complex with alpha-galactose [Bifidobacterium bifidum S17],4UZS_A Crystal structure of Bifidobacterium bifidum beta-galactosidase [Bifidobacterium bifidum S17],4UZS_B Crystal structure of Bifidobacterium bifidum beta-galactosidase [Bifidobacterium bifidum S17],4UZS_C Crystal structure of Bifidobacterium bifidum beta-galactosidase [Bifidobacterium bifidum S17] |
| 3WUF_A | 1.97e-07 | 323 | 533 | 3 | 210 | Themutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) from Streptomyces sp. 9 [Streptomyces sp.],3WUG_A The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) with xylobiose from Streptomyces sp. 9 [Streptomyces sp.] |
| 3WUB_A | 2.62e-07 | 323 | 533 | 3 | 210 | Thewild type crystal structure of b-1,4-Xylanase (XynAS9) from Streptomyces sp. 9 [Streptomyces sp.],3WUE_A The wild type crystal structure of b-1,4-Xylanase (XynAS9) with xylobiose from Streptomyces sp. 9 [Streptomyces sp.] |
| 1N82_A | 4.93e-06 | 357 | 454 | 45 | 135 | Thehigh-resolution crystal structure of IXT6, a thermophilic, intracellular xylanase from G. stearothermophilus [Geobacillus stearothermophilus],1N82_B The high-resolution crystal structure of IXT6, a thermophilic, intracellular xylanase from G. stearothermophilus [Geobacillus stearothermophilus],3MUA_A Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MUA_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus] |
| 2Q8X_A | 4.93e-06 | 357 | 454 | 45 | 135 | Thehigh-resolution crystal structure of ixt6, a thermophilic, intracellular xylanase from G. stearothermophilus [unidentified],2Q8X_B The high-resolution crystal structure of ixt6, a thermophilic, intracellular xylanase from G. stearothermophilus [unidentified],3MSD_A Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MSD_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MSG_A Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MSG_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| I1S3T9 | 1.20e-08 | 361 | 523 | 74 | 219 | Endo-1,4-beta-xylanase C OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=XYLC PE=1 SV=1 |
| D4QFE7 | 2.66e-07 | 338 | 457 | 38 | 165 | Beta-galactosidase BbgII OS=Bifidobacterium bifidum OX=1681 GN=bbgII PE=3 SV=1 |
| B4XVN1 | 6.06e-07 | 287 | 533 | 16 | 248 | Endo-1,4-beta-xylanase A OS=Streptomyces sp. OX=1931 GN=xynAS9 PE=1 SV=1 |
| Q93GI5 | 5.38e-06 | 318 | 457 | 11 | 164 | Beta-galactosidase III OS=Bifidobacterium longum subsp. infantis OX=1682 GN=beta-galIII PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
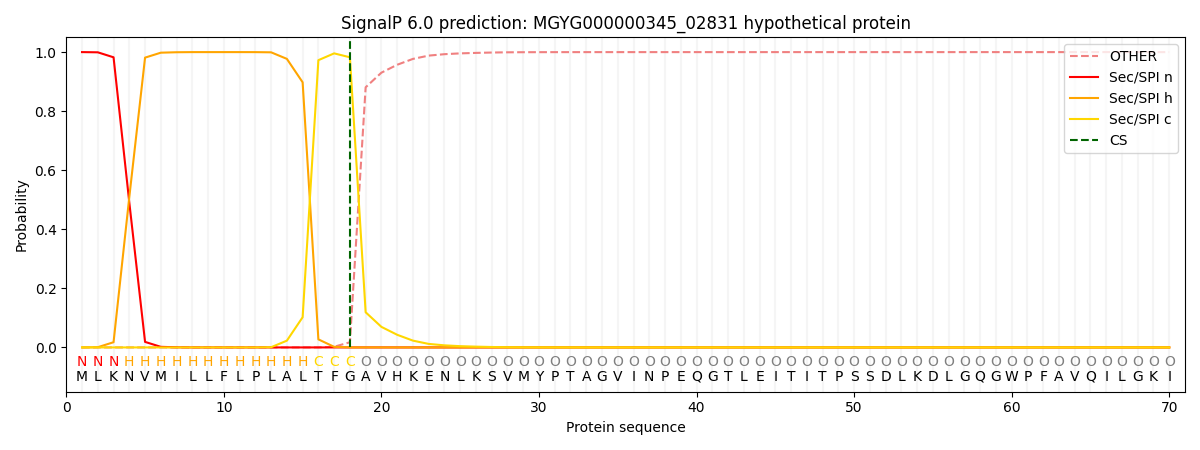
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000254 | 0.999112 | 0.000157 | 0.000163 | 0.000152 | 0.000139 |
