You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000284_00107
You are here: Home > Sequence: MGYG000000284_00107
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides sp900540715 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp900540715 | |||||||||||
| CAZyme ID | MGYG000000284_00107 | |||||||||||
| CAZy Family | GH43 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 164047; End: 165783 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH43 | 56 | 314 | 1.1e-95 | 0.9961832061068703 |
| CBM32 | 356 | 470 | 2.5e-22 | 0.8145161290322581 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd08982 | GH43-like | 1.95e-164 | 54 | 357 | 3 | 308 | Glycosyl hydrolase family 43 protein; uncharacterized. This glycosyl hydrolase family 43 (GH43)-like subfamily includes uncharacterized enzymes similar to those with beta-1,4-xylosidase (xylan 1,4-beta-xylosidase; EC 3.2.1.37), beta-1,3-xylosidase (EC 3.2.1.-), alpha-L-arabinofuranosidase (EC 3.2.1.55), arabinanase (EC 3.2.1.99), xylanase (EC 3.2.1.8), endo-alpha-L-arabinanase and galactan 1,3-beta-galactosidase (EC 3.2.1.145) activities. These are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many of the enzymes in this family display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| cd08978 | GH_F | 7.14e-22 | 57 | 286 | 1 | 245 | Glycosyl hydrolase families 43 and 62 form CAZY clan GH-F. This glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) includes family 43 (GH43) and 62 (GH62). GH43 includes enzymes with beta-xylosidase (EC 3.2.1.37), beta-1,3-xylosidase (EC 3.2.1.-), alpha-L-arabinofuranosidase (EC 3.2.1.55), arabinanase (EC 3.2.1.99), xylanase (EC 3.2.1.8), endo-alpha-L-arabinanases (beta-xylanases) and galactan 1,3-beta-galactosidase (EC 3.2.1.145) activities. GH62 includes enzymes characterized as arabinofuranosidases (alpha-L-arabinofuranosidases; EC 3.2.1.55) that specifically cleave either alpha-1,2 or alpha-1,3-L-arabinofuranose side chains from xylans. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many of the enzymes in this family display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. GH62 are also predicted to be inverting enzymes. A common structural feature of both, GH43 and GH62 enzymes, is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| cd08991 | GH43_HoAraf43-like | 2.96e-19 | 57 | 305 | 1 | 272 | Glycosyl hydrolase family 43 protein such as Halothermothrix orenii H 168 alpha-L-arabinofuranosidase (HoAraf43;Hore_20580). This glycosyl hydrolase family 43 (GH43) subgroup includes Halothermothrix orenii H 168 alpha-L-arabinofuranosidase (EC 3.2.1.55) (HoAraf43;Hore_20580). It belongs to the glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) which includes family 43 (GH43) and 62 (GH62) families. This GH43_ HoAraf43-like subgroup includes enzymes that have been annotated as having xylan-digesting beta-xylosidase (EC 3.2.1.37) and xylanase (endo-alpha-L-arabinanase, EC 3.2.1.8) activities. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| pfam00754 | F5_F8_type_C | 2.37e-18 | 352 | 478 | 1 | 124 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam04616 | Glyco_hydro_43 | 4.77e-14 | 57 | 285 | 11 | 249 | Glycosyl hydrolases family 43. The glycosyl hydrolase family 43 contains members that are arabinanases. Arabinanases hydrolyze the alpha-1,5-linked L-arabinofuranoside backbone of plant cell wall arabinans. The structure of arabinanase Arb43A from Cellvibrio japonicus reveals a five-bladed beta-propeller fold. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT49472.1 | 4.43e-199 | 248 | 578 | 15 | 345 |
| QIA09145.1 | 8.33e-168 | 34 | 574 | 35 | 575 |
| BBE16007.1 | 5.50e-167 | 29 | 574 | 26 | 570 |
| ADQ81046.1 | 7.53e-167 | 1 | 570 | 1 | 565 |
| ALM50367.1 | 3.25e-163 | 33 | 574 | 29 | 570 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4QQS_A | 2.85e-09 | 33 | 274 | 4 | 256 | Crystalstructure of a thermostable family-43 glycoside hydrolase [Halothermothrix orenii H 168],4QQS_B Crystal structure of a thermostable family-43 glycoside hydrolase [Halothermothrix orenii H 168] |
| 3BN6_A | 3.52e-06 | 333 | 465 | 15 | 141 | CrystalStructure of the C2 Domain of Bovine Lactadherin at 1.67 Angstrom Resolution [Bos taurus] |
| 2PQS_A | 3.58e-06 | 333 | 465 | 16 | 142 | CrystalStructure of the Bovine Lactadherin C2 Domain [Bos taurus],2PQS_B Crystal Structure of the Bovine Lactadherin C2 Domain [Bos taurus],2PQS_C Crystal Structure of the Bovine Lactadherin C2 Domain [Bos taurus],2PQS_D Crystal Structure of the Bovine Lactadherin C2 Domain [Bos taurus] |
| 2L9L_A | 7.86e-06 | 362 | 464 | 40 | 144 | NMRStructure of the Mouse MFG-E8 C2 Domain [Mus musculus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
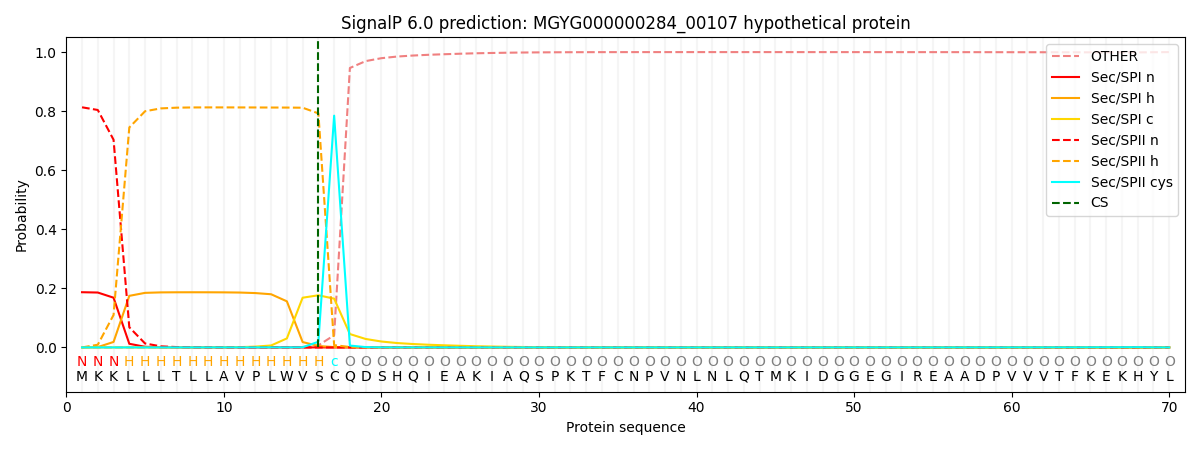
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000310 | 0.182849 | 0.816633 | 0.000083 | 0.000085 | 0.000072 |
