You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000282_01483
You are here: Home > Sequence: MGYG000000282_01483
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
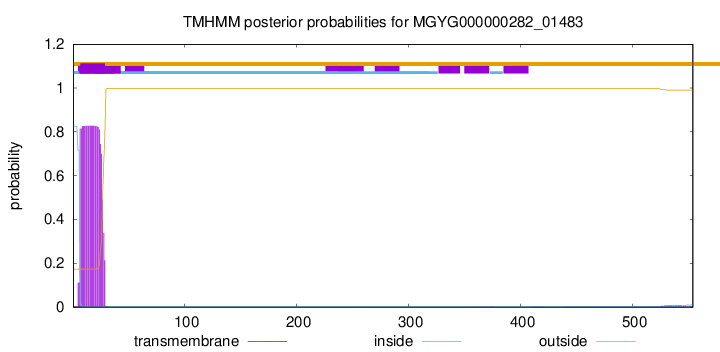
TMHMM annotations
Basic Information help
| Species | TF01-11 sp003524945 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; TF01-11; TF01-11 sp003524945 | |||||||||||
| CAZyme ID | MGYG000000282_01483 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 65671; End: 67335 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 68 | 367 | 2.2e-100 | 0.9927536231884058 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.06e-61 | 60 | 368 | 7 | 270 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 1.64e-25 | 41 | 368 | 43 | 361 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam14200 | RicinB_lectin_2 | 1.77e-14 | 415 | 494 | 14 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 0.010 | 450 | 508 | 1 | 56 | Ricin-type beta-trefoil lectin domain-like. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QYR19601.1 | 4.32e-174 | 32 | 454 | 33 | 441 |
| QAA35398.1 | 5.15e-173 | 35 | 393 | 37 | 399 |
| QYR21757.1 | 4.87e-172 | 1 | 454 | 1 | 447 |
| ADL50682.1 | 5.51e-172 | 8 | 393 | 9 | 398 |
| BAV13070.1 | 6.81e-172 | 8 | 393 | 15 | 404 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2JEP_A | 3.62e-169 | 8 | 393 | 14 | 390 | Nativefamily 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEP_B Native family 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEQ_A Family 5 xyloglucanase from Paenibacillus pabuli in complex with ligand [Paenibacillus pabuli] |
| 6WQY_A | 2.91e-78 | 44 | 382 | 26 | 365 | ChainA, Cellulase [Phocaeicola salanitronis DSM 18170] |
| 6PZ7_A | 5.23e-69 | 39 | 393 | 6 | 333 | GH5-4broad specificity endoglucanase from Clostridium acetobutylicum [Clostridium acetobutylicum ATCC 824] |
| 6WQP_A | 9.71e-68 | 39 | 393 | 13 | 352 | GH5-4broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQP_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQV_A GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_C GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_D GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis] |
| 6MQ4_A | 2.78e-66 | 39 | 393 | 9 | 345 | ChainA, cellulase [Acetivibrio cellulolyticus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O08342 | 4.10e-164 | 3 | 393 | 5 | 395 | Endoglucanase A OS=Paenibacillus barcinonensis OX=198119 GN=celA PE=1 SV=1 |
| P28621 | 3.22e-63 | 39 | 392 | 39 | 369 | Endoglucanase B OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engB PE=3 SV=1 |
| P28623 | 2.65e-62 | 39 | 392 | 40 | 367 | Endoglucanase D OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engD PE=1 SV=2 |
| P17901 | 1.53e-61 | 52 | 393 | 57 | 396 | Endoglucanase A OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCA PE=1 SV=1 |
| P54937 | 2.04e-61 | 8 | 381 | 10 | 362 | Endoglucanase A OS=Clostridium longisporum OX=1523 GN=celA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
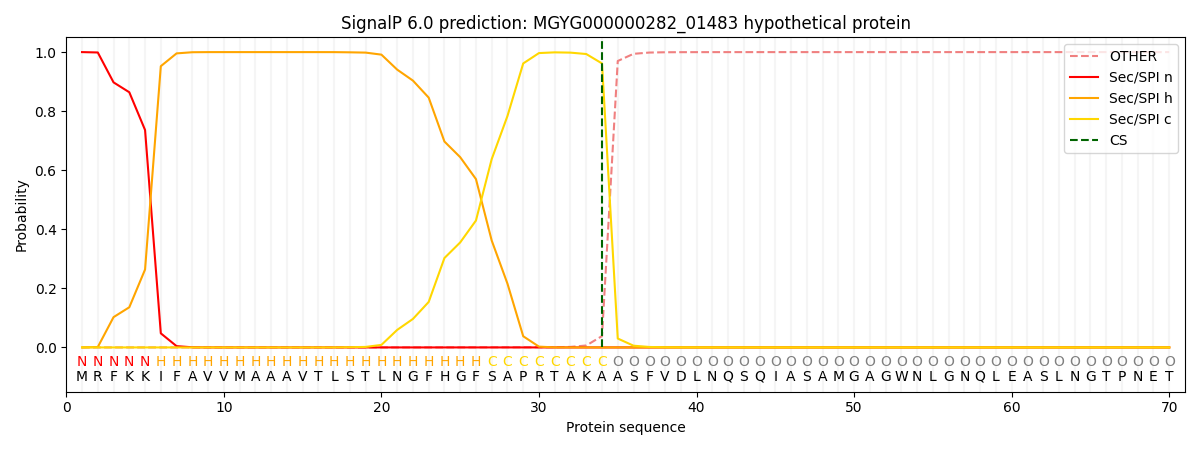
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000232 | 0.999135 | 0.000189 | 0.000166 | 0.000144 | 0.000126 |