You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000147_03404
You are here: Home > Sequence: MGYG000000147_03404
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacillus paralicheniformis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus; Bacillus paralicheniformis | |||||||||||
| CAZyme ID | MGYG000000147_03404 | |||||||||||
| CAZy Family | GH26 | |||||||||||
| CAZyme Description | Mannan endo-1,4-beta-mannosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 81146; End: 82228 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 30 | 347 | 1e-89 | 0.9933993399339934 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02156 | Glyco_hydro_26 | 1.54e-138 | 30 | 348 | 1 | 311 | Glycosyl hydrolase family 26. |
| COG4124 | ManB2 | 1.78e-94 | 2 | 360 | 6 | 355 | Beta-mannanase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QFY39913.1 | 3.79e-276 | 1 | 360 | 1 | 360 |
| AYQ15323.1 | 3.79e-276 | 1 | 360 | 1 | 360 |
| ACX94049.1 | 3.79e-276 | 1 | 360 | 1 | 360 |
| AGN35218.1 | 3.79e-276 | 1 | 360 | 1 | 360 |
| ARA84663.1 | 2.19e-275 | 1 | 360 | 1 | 360 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2QHA_A | 3.57e-214 | 25 | 360 | 1 | 336 | FromStructure to Function: Insights into the Catalytic Substrate Specificity and Thermostability Displayed by Bacillus subtilis mannanase BCman [Bacillus subtilis],2QHA_B From Structure to Function: Insights into the Catalytic Substrate Specificity and Thermostability Displayed by Bacillus subtilis mannanase BCman [Bacillus subtilis] |
| 3CBW_A | 3.77e-214 | 17 | 360 | 2 | 345 | ChainA, YdhT protein [Bacillus subtilis],3CBW_B Chain B, YdhT protein [Bacillus subtilis] |
| 2WHK_A | 3.88e-212 | 25 | 360 | 1 | 336 | Structureof Bacillus subtilis mannanase man26 [Bacillus subtilis] |
| 7EET_A | 4.87e-121 | 26 | 349 | 9 | 345 | ChainA, Mannanase KMAN [Klebsiella oxytoca] |
| 6HF2_A | 4.44e-29 | 68 | 296 | 78 | 297 | Thestructure of BoMan26B, a GH26 beta-mannanase from Bacteroides ovatus [Bacteroides ovatus ATCC 8483],6HF4_A The structure of BoMan26B, a GH26 beta-mannanase from Bacteroides ovatus, complexed with G1M4 [Bacteroides ovatus ATCC 8483] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O05512 | 8.62e-215 | 2 | 360 | 3 | 362 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis (strain 168) OX=224308 GN=gmuG PE=1 SV=2 |
| P55278 | 6.38e-198 | 1 | 360 | 2 | 360 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis OX=1423 PE=3 SV=1 |
| P16699 | 5.20e-115 | 6 | 349 | 7 | 355 | Mannan endo-1,4-beta-mannosidase A and B OS=Caldalkalibacillus mannanilyticus (strain DSM 16130 / CIP 109019 / JCM 10596 / AM-001) OX=1236954 PE=1 SV=1 |
| A1A278 | 1.79e-23 | 13 | 347 | 24 | 376 | Mannan endo-1,4-beta-mannosidase OS=Bifidobacterium adolescentis (strain ATCC 15703 / DSM 20083 / NCTC 11814 / E194a) OX=367928 GN=BAD_1030 PE=1 SV=1 |
| Q5AWB7 | 1.82e-19 | 26 | 292 | 25 | 298 | Probable mannan endo-1,4-beta-mannosidase E OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manE PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
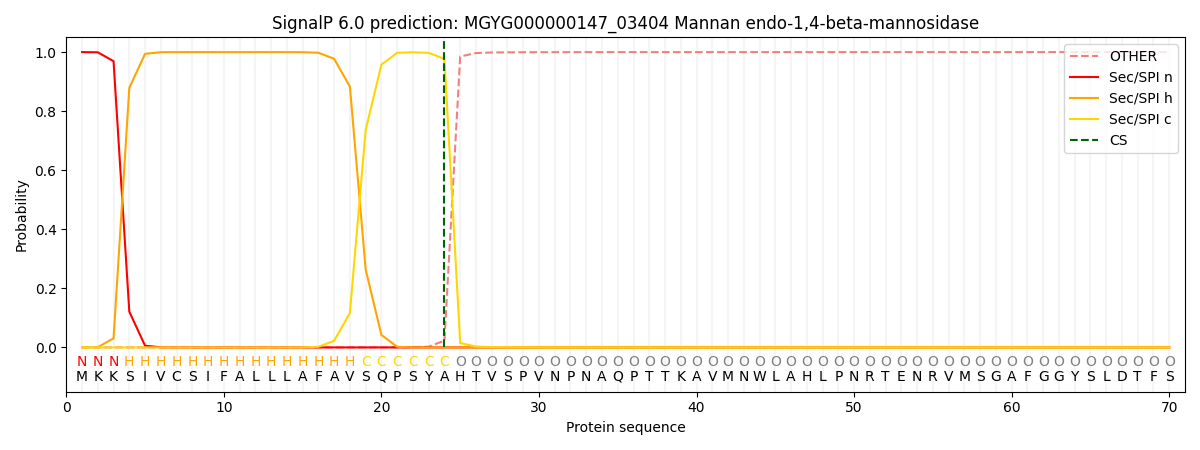
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000261 | 0.999023 | 0.000186 | 0.000168 | 0.000161 | 0.000149 |
