You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000134_00129
You are here: Home > Sequence: MGYG000000134_00129
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Longibaculum muris | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelatoclostridiaceae; Longibaculum; Longibaculum muris | |||||||||||
| CAZyme ID | MGYG000000134_00129 | |||||||||||
| CAZy Family | GH84 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 139231; End: 144927 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH84 | 179 | 478 | 3.3e-97 | 0.9627118644067797 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07555 | NAGidase | 4.87e-101 | 179 | 486 | 1 | 293 | beta-N-acetylglucosaminidase. This family has previously been described as a hyaluronidase. However, more recently it has been shown that this family has beta-N-acetylglucosaminidase activity. |
| pfam02838 | Glyco_hydro_20b | 1.66e-16 | 30 | 172 | 1 | 123 | Glycosyl hydrolase family 20, domain 2. This domain has a zincin-like fold. |
| pfam00754 | F5_F8_type_C | 2.52e-08 | 1212 | 1338 | 16 | 122 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 8.44e-08 | 950 | 1050 | 12 | 111 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| TIGR01612 | 235kDa-fam | 6.99e-06 | 1447 | 1829 | 819 | 1240 | reticulocyte binding/rhoptry protein. This model represents a group of paralogous families in plasmodium species alternately annotated as reticulocyte binding protein, 235-kDa family protein and rhoptry protein. Rhoptry protein is localized on the cell surface and is extremely large (although apparently lacking in repeat structure) and is important for the process of invasion of the RBCs by the parasite. These proteins are found in P. falciparum, P. vivax and P. yoelii. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QMW73896.1 | 0.0 | 24 | 1898 | 46 | 1892 |
| QQY28346.1 | 0.0 | 24 | 1898 | 46 | 1892 |
| QPS12837.1 | 0.0 | 24 | 1898 | 46 | 1892 |
| QQV04685.1 | 0.0 | 24 | 1898 | 46 | 1892 |
| BCL58377.1 | 0.0 | 24 | 1819 | 28 | 1822 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6PV4_A | 4.65e-241 | 31 | 642 | 32 | 653 | Structureof CpGH84A [Clostridium perfringens ATCC 13124],6PV4_B Structure of CpGH84A [Clostridium perfringens ATCC 13124],6PV4_C Structure of CpGH84A [Clostridium perfringens ATCC 13124],6PV4_D Structure of CpGH84A [Clostridium perfringens ATCC 13124] |
| 6PWI_A | 3.45e-123 | 30 | 633 | 33 | 624 | Structureof CpGH84D [Clostridium perfringens ATCC 13124],6PWI_B Structure of CpGH84D [Clostridium perfringens ATCC 13124] |
| 6PV5_A | 6.64e-54 | 33 | 554 | 42 | 554 | Structureof CpGH84B [Clostridium perfringens ATCC 13124] |
| 2XPK_A | 5.94e-53 | 35 | 544 | 18 | 512 | Cell-penetrant,nanomolar O-GlcNAcase inhibitors selective against lysosomal hexosaminidases [Clostridium perfringens],2XPK_B Cell-penetrant, nanomolar O-GlcNAcase inhibitors selective against lysosomal hexosaminidases [Clostridium perfringens] |
| 7KHV_A | 2.64e-52 | 35 | 544 | 18 | 512 | CpOGAIN COMPLEX WITH LIGAND 54 [Clostridium perfringens],7KHV_B CpOGA IN COMPLEX WITH LIGAND 54 [Clostridium perfringens],7KHV_C CpOGA IN COMPLEX WITH LIGAND 54 [Clostridium perfringens],7KHV_D CpOGA IN COMPLEX WITH LIGAND 54 [Clostridium perfringens],7KHV_E CpOGA IN COMPLEX WITH LIGAND 54 [Clostridium perfringens],7KHV_F CpOGA IN COMPLEX WITH LIGAND 54 [Clostridium perfringens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P26831 | 0.0 | 31 | 1691 | 39 | 1582 | Hyaluronoglucosaminidase OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagH PE=1 SV=2 |
| Q0TR53 | 4.15e-49 | 35 | 544 | 48 | 542 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| Q8XL08 | 2.94e-48 | 35 | 544 | 48 | 542 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
| Q89ZI2 | 3.34e-48 | 33 | 564 | 26 | 526 | O-GlcNAcase BT_4395 OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=BT_4395 PE=1 SV=1 |
| O60502 | 3.31e-19 | 180 | 437 | 63 | 321 | Protein O-GlcNAcase OS=Homo sapiens OX=9606 GN=OGA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
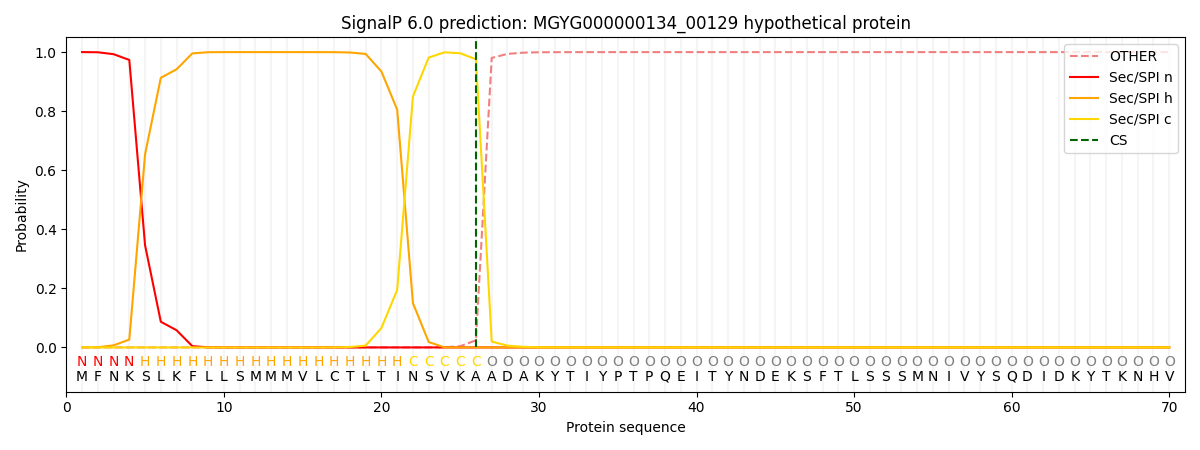
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000279 | 0.998893 | 0.000323 | 0.000174 | 0.000157 | 0.000146 |

