You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000079_02906
You are here: Home > Sequence: MGYG000000079_02906
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Exiguobacterium sp902362975 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Exiguobacterales; Exiguobacteraceae; Exiguobacterium; Exiguobacterium sp902362975 | |||||||||||
| CAZyme ID | MGYG000000079_02906 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | Cell wall-binding protein YocH | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2258; End: 3046 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3584 | 3D | 9.84e-48 | 151 | 260 | 1 | 109 | 3D (Asp-Asp-Asp) domain [Function unknown]. |
| cd14667 | 3D_containing_proteins | 7.08e-39 | 172 | 261 | 1 | 90 | Non-mltA associated 3D domain containing proteins, named for 3 conserved aspartate residues. This family contains the 3D domain, named for its 3 conserved aspartates, including similar uncharacterized proteins. These proteins contain the critical active site aspartate of mltA-like lytic transglycosylases where the 3D domain forms a larger domain with the N-terminal region. This domain is also found in conjunction with numerous other domains such as the Escherichia coli MltA, a membrane-bound lytic transglycosylase comprised of 2 domains separated by a large groove, where the peptidoglycan strand binds. Domain A is made up of an N-terminal and a C-terminal portion, corresponding to the 3D domain and Domain B is inserted within the linear sequence of domain A. MltA is distinct from other bacterial LTs, which are similar to each other. Escherichia coli peptidoglycan lytic transglycosylase (LT) initiates cell wall recycling in response to damage, during bacterial fission, and cleaves peptidoglycan (PG) to create functional spaces in its wall. PG chains (also known as murein), the major components of the bacterial cell wall, are comprised of alternating beta-1-4-linked N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), and lytic transglycosylases cleave this beta-1-4 bond. |
| PRK06347 | PRK06347 | 2.48e-23 | 23 | 172 | 328 | 484 | 1,4-beta-N-acetylmuramoylhydrolase. |
| PRK06347 | PRK06347 | 3.18e-23 | 22 | 154 | 402 | 551 | 1,4-beta-N-acetylmuramoylhydrolase. |
| pfam06725 | 3D | 7.33e-20 | 201 | 262 | 1 | 72 | 3D domain. This short presumed domain contains three conserved aspartate residues, hence the name 3D. It has been shown to be part of the catalytic double psi beta barrel domain of MltA. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUP88530.1 | 1.89e-163 | 1 | 262 | 1 | 262 |
| ACQ69197.1 | 1.89e-163 | 1 | 262 | 1 | 262 |
| QPI69077.1 | 2.11e-160 | 1 | 262 | 1 | 262 |
| QUE85347.1 | 1.43e-146 | 1 | 262 | 1 | 260 |
| ANU16064.1 | 8.78e-98 | 1 | 262 | 1 | 268 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2MKX_A | 1.27e-10 | 85 | 126 | 7 | 48 | Solutionstructure of LysM the peptidoglycan binding domain of autolysin AtlA from Enterococcus faecalis [Enterococcus faecalis V583] |
| 4WLI_A | 2.48e-09 | 165 | 261 | 18 | 113 | StationaryPhase Survival Protein YuiC from B.subtilis [Bacillus subtilis subsp. subtilis str. 168] |
| 4WLK_A | 3.54e-09 | 165 | 261 | 39 | 134 | StationaryPhase Survival Protein YuiC from B.subtilis complexed with reaction product [Bacillus subtilis subsp. subtilis str. 168],4WLK_B Stationary Phase Survival Protein YuiC from B.subtilis complexed with reaction product [Bacillus subtilis subsp. subtilis str. 168] |
| 4WJT_A | 3.60e-09 | 165 | 261 | 39 | 134 | StationaryPhase Survival Protein YuiC from B.subtilis complexed with NAG [Bacillus subtilis subsp. subtilis str. 168],4WJT_B Stationary Phase Survival Protein YuiC from B.subtilis complexed with NAG [Bacillus subtilis subsp. subtilis str. 168] |
| 4UZ2_A | 6.88e-06 | 84 | 122 | 4 | 42 | Crystalstructure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8],4UZ2_B Crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8],4UZ2_C Crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8],4UZ2_D Crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8],4UZ3_A Crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus bound to N-acetyl-chitohexaose [Thermus thermophilus HB8],4UZ3_B Crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus bound to N-acetyl-chitohexaose [Thermus thermophilus HB8],4UZ3_C Crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus bound to N-acetyl-chitohexaose [Thermus thermophilus HB8] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34669 | 8.18e-57 | 1 | 262 | 1 | 287 | Cell wall-binding protein YocH OS=Bacillus subtilis (strain 168) OX=224308 GN=yocH PE=1 SV=1 |
| P37546 | 1.71e-34 | 165 | 262 | 310 | 407 | Putative cell wall shaping protein YabE OS=Bacillus subtilis (strain 168) OX=224308 GN=yabE PE=2 SV=2 |
| Q8CMN2 | 9.60e-24 | 2 | 147 | 3 | 148 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=sle1 PE=3 SV=1 |
| Q5HRU2 | 9.60e-24 | 2 | 147 | 3 | 148 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=sle1 PE=3 SV=1 |
| Q4L3C1 | 1.94e-22 | 14 | 128 | 16 | 133 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus haemolyticus (strain JCSC1435) OX=279808 GN=sle1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
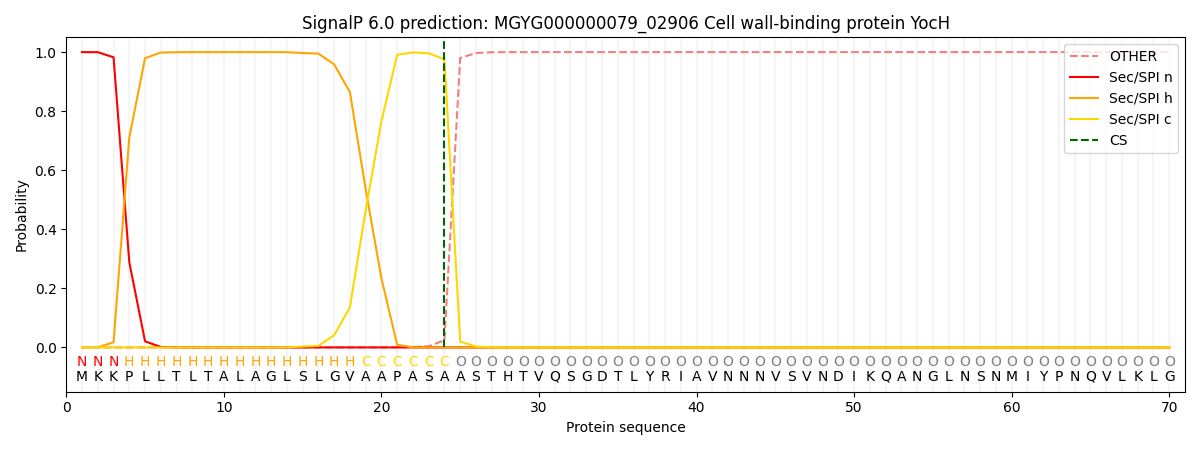
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000254 | 0.998954 | 0.000178 | 0.000220 | 0.000195 | 0.000170 |
