You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000067_01529
You are here: Home > Sequence: MGYG000000067_01529
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
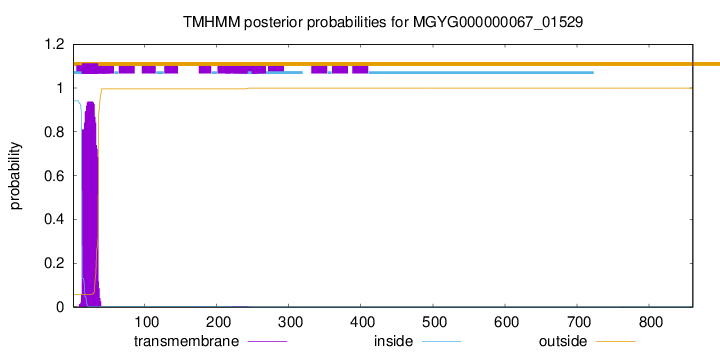
TMHMM annotations
Basic Information help
| Species | Lachnoclostridium_B sp900066765 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnoclostridium_B; Lachnoclostridium_B sp900066765 | |||||||||||
| CAZyme ID | MGYG000000067_01529 | |||||||||||
| CAZy Family | CBM13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 106416; End: 109001 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam05426 | Alginate_lyase | 4.50e-13 | 124 | 333 | 57 | 272 | Alginate lyase. This family contains several bacterial alginate lyase proteins. Alginate is a family of 1-4-linked copolymers of beta -D-mannuronic acid (M) and alpha -L-guluronic acid (G). It is produced by brown algae and by some bacteria belonging to the genera Azotobacter and Pseudomonas. Alginate lyases catalyze the depolymerization of alginates by beta -elimination, generating a molecule containing 4-deoxy-L-erythro-hex-4-enepyranosyluronate at the nonreducing end. This family adopts an all alpha fold. |
| pfam14200 | RicinB_lectin_2 | 9.50e-13 | 468 | 552 | 5 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam00395 | SLH | 9.02e-08 | 736 | 778 | 1 | 42 | S-layer homology domain. |
| pfam07554 | FIVAR | 9.67e-07 | 574 | 635 | 1 | 68 | FIVAR domain. This domain is found in a wide variety of contexts, but mostly occurring in cell wall associated proteins. A lack of conserved catalytic residues suggests that it is a binding domain. From context, possible substrates are hyaluronate or fibronectin (personal obs: C Yeats). This is further evidenced by. Possibly the exact substrate is N-acetyl glucosamine. Finding it in the same protein as pfam05089 further supports this proposal. It is found in the C-terminal part of Bacillus sp. Gellan lyase, which is removed during maturation. Some of the proteins it is found in are involved in methicillin resistance. The name FIVAR derives from Found In Various Architectures. |
| NF033190 | inl_like_NEAT_1 | 1.09e-06 | 676 | 834 | 582 | 734 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDA29257.1 | 1.23e-154 | 58 | 566 | 35 | 555 |
| AHM66085.1 | 4.85e-154 | 58 | 566 | 35 | 555 |
| AIY11542.1 | 4.85e-154 | 58 | 566 | 35 | 555 |
| QPK60353.1 | 6.85e-154 | 58 | 566 | 35 | 555 |
| QPK55271.1 | 6.85e-154 | 58 | 566 | 35 | 555 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4G9M_A | 7.76e-06 | 434 | 562 | 8 | 137 | Crystalstructure of the Rhizoctonia solani agglutinin [Rhizoctonia solani],4G9M_B Crystal structure of the Rhizoctonia solani agglutinin [Rhizoctonia solani],4G9N_A Crystal structure of the Rhizoctonia solani agglutinin in complex with N'-acetyl-galactosamine [Rhizoctonia solani],4G9N_B Crystal structure of the Rhizoctonia solani agglutinin in complex with N'-acetyl-galactosamine [Rhizoctonia solani] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P19424 | 1.97e-15 | 675 | 853 | 40 | 214 | Endoglucanase OS=Bacillus sp. (strain KSM-635) OX=1415 PE=1 SV=1 |
| C6CRV0 | 3.21e-13 | 676 | 851 | 1283 | 1457 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
| P38537 | 2.12e-08 | 696 | 853 | 54 | 206 | Surface-layer 125 kDa protein OS=Lysinibacillus sphaericus OX=1421 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
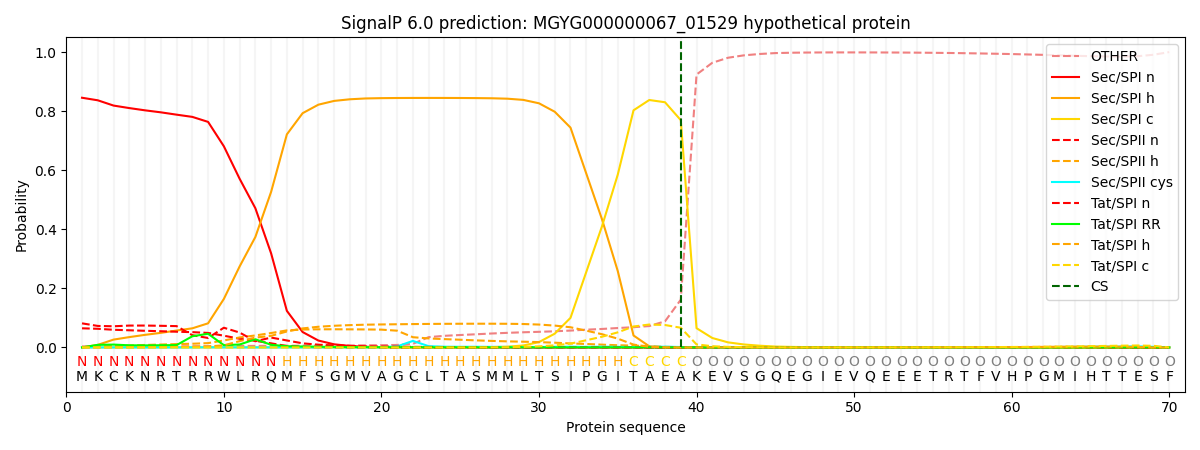
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.009034 | 0.819328 | 0.077591 | 0.086469 | 0.006649 | 0.000910 |