You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000047_00882
You are here: Home > Sequence: MGYG000000047_00882
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Clostridium disporicum_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium; Clostridium disporicum_A | |||||||||||
| CAZyme ID | MGYG000000047_00882 | |||||||||||
| CAZy Family | CBM51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 25027; End: 31878 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM51 | 1826 | 1965 | 3.4e-43 | 0.9850746268656716 |
| CBM51 | 1311 | 1445 | 2.9e-35 | 0.9850746268656716 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08305 | NPCBM | 2.86e-51 | 1823 | 1966 | 1 | 136 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 6.25e-43 | 1825 | 1966 | 5 | 145 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| pfam08305 | NPCBM | 1.88e-42 | 1308 | 1446 | 1 | 136 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 1.45e-33 | 1311 | 1446 | 6 | 145 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| pfam16403 | DUF5011 | 2.98e-15 | 1760 | 1818 | 9 | 71 | Domain of unknown function (DUF5011). This small family of proteins is functionally uncharacterized. This family is found in Bacteroides, Prevotella, and Parabateroides. Proteins in this family are around 230 amino acids in length. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJS19250.1 | 0.0 | 2 | 2280 | 1 | 2286 |
| AQM59432.1 | 0.0 | 3 | 2280 | 3 | 2441 |
| CED93629.1 | 0.0 | 3 | 2283 | 3 | 1923 |
| AIY84110.1 | 0.0 | 3 | 2280 | 3 | 1921 |
| ABG84962.1 | 0.0 | 2 | 2276 | 1 | 2135 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7JFS_A | 2.03e-188 | 47 | 1229 | 26 | 981 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 7JS4_A | 2.74e-131 | 723 | 1512 | 28 | 819 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 7JND_A | 3.25e-127 | 28 | 453 | 27 | 449 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124],7JNF_A Chain A, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 7JNB_A | 1.33e-125 | 28 | 453 | 27 | 449 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 5KDJ_A | 1.52e-95 | 655 | 1307 | 24 | 672 | ZmpBmetallopeptidase from Clostridium perfringens [Clostridium perfringens ATCC 13124],5KDJ_B ZmpB metallopeptidase from Clostridium perfringens [Clostridium perfringens ATCC 13124] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
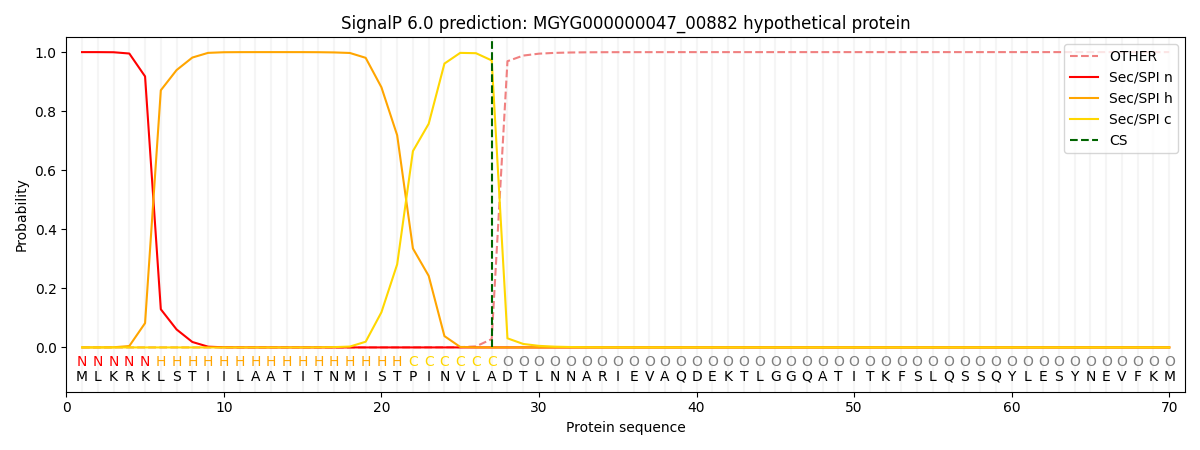
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000278 | 0.999013 | 0.000179 | 0.000195 | 0.000163 | 0.000140 |
