You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000014_01755
You are here: Home > Sequence: MGYG000000014_01755
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
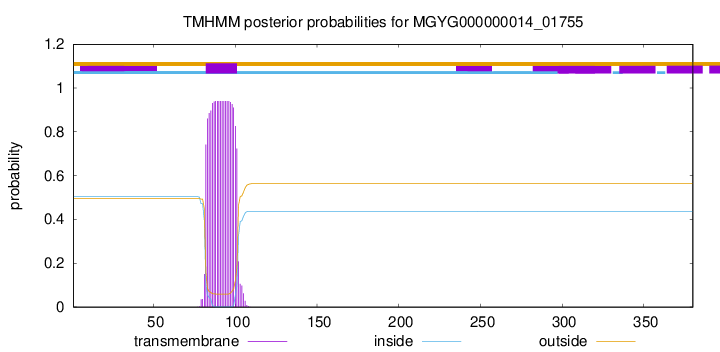
TMHMM annotations
Basic Information help
| Species | Clostridium butyricum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium; Clostridium butyricum | |||||||||||
| CAZyme ID | MGYG000000014_01755 | |||||||||||
| CAZy Family | CE4 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 30662; End: 31804 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE4 | 176 | 311 | 3.2e-25 | 0.9076923076923077 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd10944 | CE4_SmPgdA_like | 1.85e-81 | 179 | 378 | 1 | 189 | Catalytic NodB homology domain of Streptococcus mutans polysaccharide deacetylase PgdA, Bacillus subtilis YheN, and similar proteins. This family is represented by a putative polysaccharide deacetylase PgdA from the oral pathogen Streptococcus mutans (SmPgdA) and Bacillus subtilis YheN (BsYheN), which are members of the carbohydrate esterase 4 (CE4) superfamily. SmPgdA is an extracellular metal-dependent polysaccharide deacetylase with a typical CE4 fold, with metal bound to a His-His-Asp triad. It possesses de-N-acetylase activity toward a hexamer of chitooligosaccharide N-acetylglucosamine, but not shorter chitooligosaccharides or a synthetic peptidoglycan tetrasaccharide. SmPgdA plays a role in tuning cell surface properties and in interactions with (salivary) agglutinin, an essential component of the innate immune system, most likely through deacetylation of an as-yet-unidentified polysaccharide. SmPgdA shows significant homology to the catalytic domains of peptidoglycan deacetylases from Streptococcus pneumoniae (SpPgdA) and Listeria monocytogenes (LmPgdA), both of which are involved in the bacterial defense mechanism against human mucosal lysozyme. The Bacillus subtilis genome contains six polysaccharide deacetylase gene homologs: pdaA, pdaB (previously known as ybaN), yheN, yjeA, yxkH and ylxY. The biological function of BsYheN is still unknown. This family also includes many uncharacterized polysaccharide deacetylases mainly found in bacteria. |
| cd10917 | CE4_NodB_like_6s_7s | 3.75e-49 | 179 | 370 | 1 | 171 | Catalytic NodB homology domain of rhizobial NodB-like proteins. This family belongs to the large and functionally diverse carbohydrate esterase 4 (CE4) superfamily, whose members show strong sequence similarity with some variability due to their distinct carbohydrate substrates. It includes many rhizobial NodB chitooligosaccharide N-deacetylase (EC 3.5.1.-)-like proteins, mainly from bacteria and eukaryotes, such as chitin deacetylases (EC 3.5.1.41), bacterial peptidoglycan N-acetylglucosamine deacetylases (EC 3.5.1.-), and acetylxylan esterases (EC 3.1.1.72), which catalyze the N- or O-deacetylation of substrates such as acetylated chitin, peptidoglycan, and acetylated xylan. All members of this family contain a catalytic NodB homology domain with the same overall topology and a deformed (beta/alpha)8 barrel fold with 6- or 7 strands. Their catalytic activity is dependent on the presence of a divalent cation, preferably cobalt or zinc, and they employ a conserved His-His-Asp zinc-binding triad closely associated with the conserved catalytic base (aspartic acid) and acid (histidine) to carry out acid/base catalysis. Several family members show diversity both in metal ion specificities and in the residues that coordinate the metal. |
| cd10954 | CE4_CtAXE_like | 1.09e-37 | 179 | 379 | 1 | 177 | Catalytic NodB homology domain of Clostridium thermocellum acetylxylan esterase and its bacterial homologs. This family is represented by Clostridium thermocellum acetylxylan esterase (CtAXE, EC 3.1.1.72), a member of the carbohydrate esterase 4 (CE4) superfamily. CtAXE deacetylates O-acetylated xylan, a key component of plant cell walls. It shows no detectable activity on generic esterase substrates including para-nitrophenyl acetate. It is specific for sugar-based substrates and will precipitate acetylxylan, as a consequence of deacetylation. CtAXE is a monomeric protein containing a catalytic NodB homology domain with the same overall topology and a deformed (beta/alpha)8 barrel fold as other CE4 esterases. However, due to differences in the topography of the substrate-binding groove, the chemistry of the active center, and metal ion coordination, CtAXE has different metal ion preference and lacks activity on N-acetyl substrates. It is significantly activated by Co2+. Moreover, CtAXE displays distinctly different ligand coordination to the metal ion, utilizing an aspartate, a histidine, and four water molecules, as opposed to the conserved His-His-Asp zinc-binding triad of other CE4 esterases. |
| cd10956 | CE4_BH1302_like | 1.12e-34 | 178 | 380 | 4 | 190 | Putative catalytic NodB homology domain of uncharacterized BH1302 protein from Bacillus halodurans and its bacterial homologs. This family is represented by a putative polysaccharide deacetylase BH1302 from Bacillus halodurans. Although its biological function is unknown, BH1302 shows high sequence homology to the catalytic NodB homology domain of Streptococcus pneumoniae polysaccharide deacetylase PgdA (SpPgdA), which is an extracellular metal-dependent polysaccharide deacetylase with de-N-acetylase activity toward a hexamer of chitooligosaccharide N-acetylglucosamine, but not shorter chitooligosaccharides or a synthetic peptidoglycan tetrasaccharide. Both BH1302 and SpPgdA belong to the carbohydrate esterase 4 (CE4) superfamily. This family also includes many uncharacterized bacterial polysaccharide deacetylases. |
| COG0726 | CDA1 | 6.64e-34 | 175 | 379 | 61 | 253 | Peptidoglycan/xylan/chitin deacetylase, PgdA/CDA1 family [Carbohydrate transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| APF22938.1 | 4.91e-264 | 1 | 380 | 1 | 380 |
| ALP89619.1 | 4.91e-264 | 1 | 380 | 1 | 380 |
| QCJ07029.1 | 4.91e-264 | 1 | 380 | 1 | 380 |
| QCJ01964.1 | 4.91e-264 | 1 | 380 | 1 | 380 |
| ALS16074.1 | 4.91e-264 | 1 | 380 | 1 | 380 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2W3Z_A | 6.09e-45 | 128 | 379 | 57 | 310 | Structureof a Streptococcus mutans CE4 esterase [Streptococcus mutans UA159] |
| 5JMU_A | 2.61e-27 | 164 | 380 | 5 | 214 | ChainA, Peptidoglycan N-acetylglucosamine deacetylase [[Eubacterium] rectale ATCC 33656] |
| 2C1G_A | 1.71e-26 | 178 | 379 | 235 | 412 | Structureof Streptococcus pneumoniae peptidoglycan deacetylase (SpPgdA) [Streptococcus pneumoniae R6] |
| 2C1I_A | 8.17e-26 | 178 | 379 | 235 | 412 | Structureof Streptococcus pneumoniae peptidoglycan deacetylase (SpPgdA) D 275 N Mutant. [Streptococcus pneumoniae R6] |
| 7AX7_A | 3.94e-20 | 176 | 380 | 1 | 183 | Crystalstructure of the Xyl-CE4 domain of a multidomain xylanase from the hindgut metagenome of Trinervitermes trinervoides [uncultured bacterium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8DP63 | 1.27e-25 | 178 | 379 | 267 | 444 | Peptidoglycan-N-acetylglucosamine deacetylase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=pgdA PE=1 SV=1 |
| Q81EK9 | 2.46e-18 | 181 | 379 | 83 | 264 | Peptidoglycan-N-acetylglucosamine deacetylase BC_1960 OS=Bacillus cereus (strain ATCC 14579 / DSM 31 / CCUG 7414 / JCM 2152 / NBRC 15305 / NCIMB 9373 / NCTC 2599 / NRRL B-3711) OX=226900 GN=BC_1960 PE=1 SV=1 |
| Q81AF4 | 2.34e-16 | 178 | 380 | 21 | 206 | Peptidoglycan-N-acetylglucosamine deacetylase BC_3618 OS=Bacillus cereus (strain ATCC 14579 / DSM 31 / CCUG 7414 / JCM 2152 / NBRC 15305 / NCIMB 9373 / NCTC 2599 / NRRL B-3711) OX=226900 GN=BC_3618 PE=1 SV=1 |
| Q81EJ6 | 2.54e-16 | 175 | 377 | 64 | 255 | Peptidoglycan-N-acetylglucosamine deacetylase BC_1974 OS=Bacillus cereus (strain ATCC 14579 / DSM 31 / CCUG 7414 / JCM 2152 / NBRC 15305 / NCIMB 9373 / NCTC 2599 / NRRL B-3711) OX=226900 GN=BC_1974 PE=1 SV=1 |
| P83513 | 2.70e-16 | 178 | 380 | 401 | 581 | Bifunctional xylanase/deacetylase OS=Pseudobutyrivibrio xylanivorans OX=185007 GN=xyn11A PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
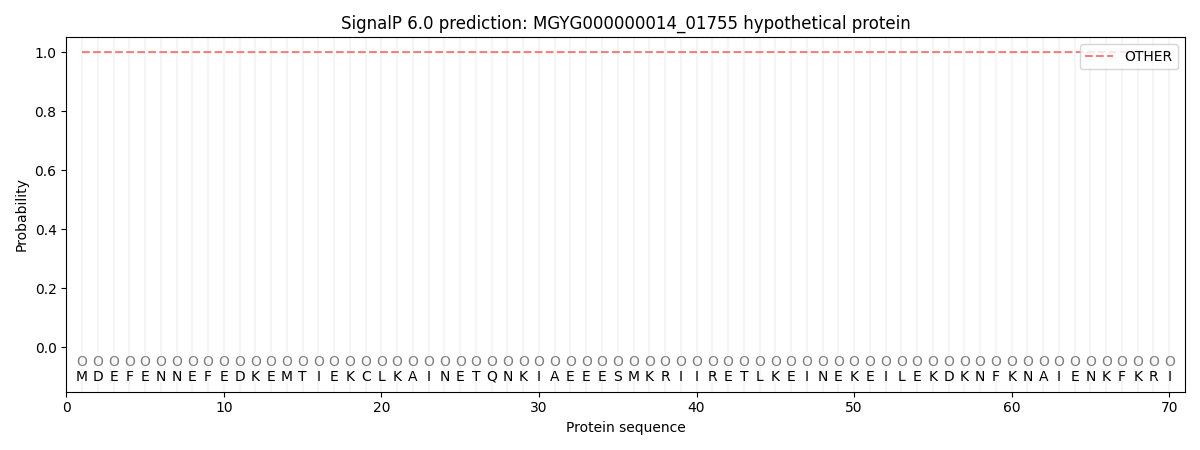
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000039 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |